SFRP2/DPP4 and FMO1/LSP1 Define Major Fibroblast Populations in Human Skin
- PMID: 29080679
- PMCID: PMC7444611
- DOI: 10.1016/j.jid.2017.09.045
SFRP2/DPP4 and FMO1/LSP1 Define Major Fibroblast Populations in Human Skin
Erratum in
-
Addendum.J Invest Dermatol. 2018 Sep;138(9):2086. doi: 10.1016/j.jid.2018.07.003. J Invest Dermatol. 2018. PMID: 30143084 No abstract available.
Abstract
Fibroblasts produce matrix, regulate inflammation, mediate reparative processes, and serve as pluripotent mesenchymal cells. Analyzing digested normal human skin by single-cell RNA sequencing, we explored different fibroblast populations. T-distributed stochastic neighbor embedding and clustering of single-cell RNA sequencing data from six biopsy samples showed two major fibroblast populations, defined by distinct genes, including SFRP2 and FMO1, expressed exclusively by these two major fibroblast populations. Further subpopulations were defined within each of the SFRP2 and FMO1 populations and five minor fibroblast populations, each expressing discrete genes: CRABP1, COL11A1, FMO2, PRG4, or C2ORF40. Immunofluorescent staining confirmed that SFRP2 and FMO1 define cell types of dramatically different morphology. SFRP2+ fibroblasts were small, elongated, and distributed between collagen bundles. FMO1+ fibroblasts were larger and distributed in both interstitial and perivascular locations. Differential gene expression by SFRP2+, FMO1+, and COL11A1+ fibroblasts suggests roles in matrix deposition, inflammatory cell retention, and connective tissue cell differentiation, respectively.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
CONFLICT OF INTEREST STATEMENT: Dr. Lafyatis has received consulting fees from Merck, Bristol Myers Squibb, Biocon, Formation, Genentech/Roche, UCB and Sanofi; and grant support from Momenta, Regeneron and PRISM. The other authors have declared that no conflict of interest exists.
Figures
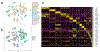



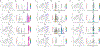

Comment in
-
A Fibroblast Is Not a Fibroblast Is Not a Fibroblast.J Invest Dermatol. 2018 Apr;138(4):729-730. doi: 10.1016/j.jid.2017.10.012. J Invest Dermatol. 2018. PMID: 29579454 Free PMC article.
References
-
- Collins CA, Watt FM. Dynamic regulation of retinoic acid-binding proteins in developing, adult and neoplastic skin reveals roles for beta-catenin and Notch signalling. Developmental biology 2008;324(1):55–67. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

