Critical assessment of methods of protein structure prediction (CASP)-Round XII
- PMID: 29082672
- PMCID: PMC5897042
- DOI: 10.1002/prot.25415
Critical assessment of methods of protein structure prediction (CASP)-Round XII
Abstract
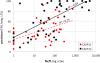
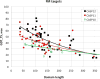
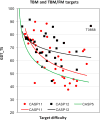
This article reports the outcome of the 12th round of Critical Assessment of Structure Prediction (CASP12), held in 2016. CASP is a community experiment to determine the state of the art in modeling protein structure from amino acid sequence. Participants are provided sequence information and in turn provide protein structure models and related information. Analysis of the submitted structures by independent assessors provides a comprehensive picture of the capabilities of current methods, and allows progress to be identified. This was again an exciting round of CASP, with significant advances in 4 areas: (i) The use of new methods for predicting three-dimensional contacts led to a two-fold improvement in contact accuracy. (ii) As a consequence, model accuracy for proteins where no template was available improved dramatically. (iii) Models based on a structural template showed overall improvement in accuracy. (iv) Methods for estimating the accuracy of a model continued to improve. CASP continued to develop new areas: (i) Assessing methods for building quaternary structure models, including an expansion of the collaboration between CASP and CAPRI. (ii) Modeling with the aid of experimental data was extended to include SAXS data, as well as again using chemical cross-linking information. (iii) A team of assessors evaluated the suitability of models for a range of applications, including mutation interpretation, analysis of ligand binding properties, and identification of interfaces. This article describes the experiment and summarizes the results. The rest of this special issue of PROTEINS contains papers describing CASP12 results and assessments in more detail.
Keywords: CASP; community wide experiment; protein structure prediction.
© 2017 Wiley Periodicals, Inc.
Figures




References
-
- Dal Peraro M, et al. CASP targets paper. PROTEINS. 2017 CASP12 issue.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

