Metagenomic analysis of viral diversity in respiratory samples from patients with respiratory tract infections in Kuwait
- PMID: 29083040
- PMCID: PMC7167075
- DOI: 10.1002/jmv.24984
Metagenomic analysis of viral diversity in respiratory samples from patients with respiratory tract infections in Kuwait
Abstract
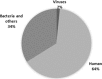
A metagenomic approach based on target independent next-generation sequencing has become a known method for the detection of both known and novel viruses in clinical samples. This study aimed to use the metagenomic sequencing approach to characterize the viral diversity in respiratory samples from patients with respiratory tract infections. We have investigated 86 respiratory samples received from various hospitals in Kuwait between 2015 and 2016 for the diagnosis of respiratory tract infections. A metagenomic approach using the next-generation sequencer to characterize viruses was used. According to the metagenomic analysis, an average of 145, 019 reads were identified, and 2% of these reads were of viral origin. Also, metagenomic analysis of the viral sequences revealed many known respiratory viruses, which were detected in 30.2% of the clinical samples. Also, sequences of non-respiratory viruses were detected in 14% of the clinical samples, while sequences of non-human viruses were detected in 55.8% of the clinical samples. The average genome coverage of the viruses was 12% with the highest genome coverage of 99.2% for respiratory syncytial virus, and the lowest was 1% for torque teno midi virus 2. Our results showed 47.7% agreement between multiplex Real-Time PCR and metagenomics sequencing in the detection of respiratory viruses in the clinical samples. Though there are some difficulties in using this method to clinical samples such as specimen quality, these observations are indicative of the promising utility of the metagenomic sequencing approach for the identification of respiratory viruses in patients with respiratory tract infections.
Keywords: RNA extraction; human metapneumovirus; influenza virus; research and analysis methods; respiratory syncytial virus.
© 2017 Wiley Periodicals, Inc.
Figures


References
-
- Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol. 1998; 5:R245–R249. - PubMed
-
- Riesenfeld CS, Schloss PD, Handelsman J. Metagenomics: genomic analysis of microbial communities. Annu Rev Genet. 2004; 38:525–552. - PubMed
-
- Krause DO, Denman SE, Mackie RI, et al. Opportunities to improve fiber degradation in the rumen: microbiology, ecology, and genomics. FEMS Microbiol Rev. 2003; 27:663–693. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

