Evaluating Metabolite-Related DNA Oxidation and Adduct Damage from Aryl Amines Using a Microfluidic ECL Array
- PMID: 29083162
- PMCID: PMC5777145
- DOI: 10.1021/acs.analchem.7b03528
Evaluating Metabolite-Related DNA Oxidation and Adduct Damage from Aryl Amines Using a Microfluidic ECL Array
Abstract
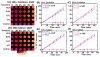
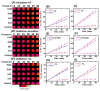
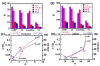
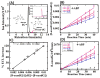
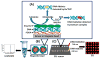
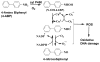
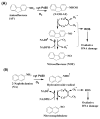
Damage to DNA from the metabolites of drugs and pollutants constitutes a major human toxicity pathway known as genotoxicity. Metabolites can react with metal ions and NADPH to oxidize DNA or participate in SN2 reactions to form covalently linked adducts with DNA bases. Guanines are the main DNA oxidation sites, and 8-oxo-7,8-dihydro-2-deoxyguanosine (8-oxodG) is the initial product. Here we describe a novel electrochemiluminescent (ECL) microwell array that produces metabolites from test compounds and measures relative rates of DNA oxidation and DNA adduct damage. In this new array, films of DNA, metabolic enzymes, and an ECL metallopolymer or complex assembled in microwells on a pyrolytic graphite wafer are housed in dual microfluidic chambers. As reactant solution passes over the wells, metabolites form and can react with DNA in the films to form DNA adducts. These adducts are detected by ECL from a RuPVP polymer that uses DNA as a coreactant. Aryl amines also combine with Cu2+ and NADPH to form reactive oxygen species (ROS) that oxidize DNA. The resulting 8-oxodG was detected selectively by ECL-generating bis(2,2'-bipyridine)-(4-(1,10-phenanthrolin-6-yl)-benzoic acid)Os(II). DNA/enzyme films on magnetic beads were oxidized similarly, and 8-oxodG determined by LC/MS/MS enabled array standardization. The array limit of detection for oxidation was 720 8-oxodG per 106 nucleobases. For a series of aryl amines, metabolite-generated DNA oxidation and adduct formation turnover rates from the array correlated very well with rodent 1/TD50 and Comet assay results.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
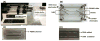
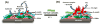
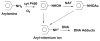

Similar articles
-
Metabolites of Tobacco- and E-Cigarette-Related Nitrosamines Can Drive Cu2+-Mediated DNA Oxidation.Chem Res Toxicol. 2020 Aug 17;33(8):2072-2086. doi: 10.1021/acs.chemrestox.0c00027. Epub 2020 Aug 4. Chem Res Toxicol. 2020. PMID: 32672941 Free PMC article.
-
Electrochemiluminescent Array to Detect Oxidative Damage in ds-DNA Using [Os(bpy)2(phen-benz-COOH)]2+/Nafion/Graphene Films.ACS Sens. 2016;1(3):272-278. doi: 10.1021/acssensors.5b00189. Epub 2016 Jan 8. ACS Sens. 2016. PMID: 27135053 Free PMC article.
-
High-throughput metabolic genotoxicity screening with a fluidic microwell chip and electrochemiluminescence.Lab Chip. 2013 Dec 7;13(23):4554-62. doi: 10.1039/c3lc50698c. Lab Chip. 2013. PMID: 24113555 Free PMC article.
-
Screening Genotoxicity Chemistry with Microfluidic Electrochemiluminescent Arrays.Sensors (Basel). 2017 May 3;17(5):1008. doi: 10.3390/s17051008. Sensors (Basel). 2017. PMID: 28467352 Free PMC article. Review.
-
Mechanisms of oxidative DNA damage induced by carcinogenic arylamines.Front Biosci (Landmark Ed). 2011 Jan 1;16(3):1132-43. doi: 10.2741/3739. Front Biosci (Landmark Ed). 2011. PMID: 21196222 Review.
Cited by
-
Metabolites of Tobacco- and E-Cigarette-Related Nitrosamines Can Drive Cu2+-Mediated DNA Oxidation.Chem Res Toxicol. 2020 Aug 17;33(8):2072-2086. doi: 10.1021/acs.chemrestox.0c00027. Epub 2020 Aug 4. Chem Res Toxicol. 2020. PMID: 32672941 Free PMC article.
-
Organ-Specific Screening for Protein Damage Using Magnetic Bead Bioreactors and LC-MS/MS.Anal Chem. 2020 Apr 7;92(7):5337-5345. doi: 10.1021/acs.analchem.9b05871. Epub 2020 Mar 24. Anal Chem. 2020. PMID: 32176468 Free PMC article.
-
Highly sensitive detection of MMP-2 using an electrochemiluminescent biosensor enhanced by ladder-branch hybridization chain reaction.Mikrochim Acta. 2024 Nov 13;191(12):742. doi: 10.1007/s00604-024-06761-y. Mikrochim Acta. 2024. PMID: 39535619
-
Confined Electrochemiluminescence Generation at Ultra-High-Density Gold Microwell Electrodes.Front Chem. 2021 Jan 26;8:630246. doi: 10.3389/fchem.2020.630246. eCollection 2020. Front Chem. 2021. PMID: 33575249 Free PMC article.
References
-
- Park BK, Boobis A, Clarke S, Goldring CEP, Jones D, Kenna JG, Lambert C, Laverty HG, Naisbitt DJ, Nelson S, Nicoll-Griffith DA, Obach RS, Routledge P, Smith DA, Tweedie DJ, Vermeulen N, Williams DP, Wilson ID, Baillie TA. Nat Rev Drug Discovery. 2011;10:292–307. - PubMed
-
- Farmer PB, Brown K, Tompkins E, Emms VL, Jones DJ, Singh R, Phillips DH. Toxicol Appl Pharmacol. 2005;207:293–301. - PubMed
-
- Spencer WA, Vadhanam MV, Jeyabalan J, Gupta RC. Chem Res Toxicol. 2012;25:305–314. - PubMed
-
- Murata M, Kawanishi S Front. Biosci, Landmark Ed. 2011;16:1132–1143. - PubMed
-
- Bowes J, Brown AJ, Hamon J, Jarolimek W, Sridhar A, Waldron G, Whitebread S. Nat Rev Drug Discovery. 2012;11:909–922. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

