Ancient hybridization and strong adaptation to viruses across African vervet monkey populations
- PMID: 29083404
- PMCID: PMC5709169
- DOI: 10.1038/ng.3980
Ancient hybridization and strong adaptation to viruses across African vervet monkey populations
Erratum in
-
Publisher Correction: Ancient hybridization and strong adaptation to viruses across African vervet monkey populations.Nat Genet. 2018 Nov;50(11):1617. doi: 10.1038/s41588-018-0124-x. Nat Genet. 2018. PMID: 30327573
Abstract
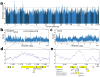
Vervet monkeys are among the most widely distributed nonhuman primates, show considerable phenotypic diversity, and have long been an important biomedical model for a variety of human diseases and in vaccine research. Using whole-genome sequencing data from 163 vervets sampled from across Africa and the Caribbean, we find high diversity within and between taxa and clear evidence that taxonomic divergence was reticulate rather than following a simple branching pattern. A scan for diversifying selection across taxa identifies strong and highly polygenic selection signals affecting viral processes. Furthermore, selection scores are elevated in genes whose human orthologs interact with HIV and in genes that show a response to experimental simian immunodeficiency virus (SIV) infection in vervet monkeys but not in rhesus macaques, suggesting that part of the signal reflects taxon-specific adaptation to SIV.
Conflict of interest statement
The authors declare that there are no competing financial interests.
Figures

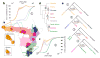


Comment in
-
Evolutionary insights from wild vervet genomes.Nat Genet. 2017 Nov 29;49(12):1671-1672. doi: 10.1038/ng.3992. Nat Genet. 2017. PMID: 29186128 Free PMC article.
References
-
- Cheney DL, Seyfarth RM. The recognition of social alliances by vervet monkeys. Anim Behav. 1986;34:1722–1731.
-
- Seyfarth RM, Cheney DL, Marler P. Vervet monkey alarm calls: Semantic communication in a free-ranging primate. Anim Behav. 1980;28:1070–1094.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

