Genetic variation and gene expression across multiple tissues and developmental stages in a nonhuman primate
- PMID: 29083405
- PMCID: PMC5714271
- DOI: 10.1038/ng.3959
Genetic variation and gene expression across multiple tissues and developmental stages in a nonhuman primate
Abstract
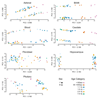
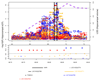
By analyzing multitissue gene expression and genome-wide genetic variation data in samples from a vervet monkey pedigree, we generated a transcriptome resource and produced the first catalog of expression quantitative trait loci (eQTLs) in a nonhuman primate model. This catalog contains more genome-wide significant eQTLs per sample than comparable human resources and identifies sex- and age-related expression patterns. Findings include a master regulatory locus that likely has a role in immune function and a locus regulating hippocampal long noncoding RNAs (lncRNAs), whose expression correlates with hippocampal volume. This resource will facilitate genetic investigation of quantitative traits, including brain and behavioral phenotypes relevant to neuropsychiatric disorders.
Figures
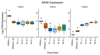


References
-
- Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nat Rev Genet. 2015;16:197–212. - PubMed
MeSH terms
Grants and funding
- R37 MH057881/MH/NIMH NIH HHS/United States
- T32 MH073526/MH/NIMH NIH HHS/United States
- P40 RR019963/RR/NCRR NIH HHS/United States
- RL1 MH083270/MH/NIMH NIH HHS/United States
- UL1 DE019580/DE/NIDCR NIH HHS/United States
- P30 NS062691/NS/NINDS NIH HHS/United States
- R37 MH060233/MH/NIMH NIH HHS/United States
- R01 RR016300/RR/NCRR NIH HHS/United States
- PL1 NS062410/NS/NINDS NIH HHS/United States
- P50 MH084053/MH/NIMH NIH HHS/United States
- 108749/WT_/Wellcome Trust/United Kingdom
- HHSN271201300031C/MH/NIMH NIH HHS/United States
- P50 MH066392/MH/NIMH NIH HHS/United States
- R01 MH080405/MH/NIMH NIH HHS/United States
- R01 MH085542/MH/NIMH NIH HHS/United States
- P40 OD010965/OD/NIH HHS/United States
- R01 OD010980/OD/NIH HHS/United States
- R01 MH101782/MH/NIMH NIH HHS/United States
- R01 MH075916/MH/NIMH NIH HHS/United States
- R01 MH097276/MH/NIMH NIH HHS/United States
- P01 AG002219/AG/NIA NIH HHS/United States
- R01 MH093725/MH/NIMH NIH HHS/United States
- P50 AG005138/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

