Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology
- PMID: 29083406
- PMCID: PMC5989923
- DOI: 10.1038/ng.3985
Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology
Abstract
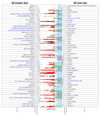
Asthma, hay fever (or allergic rhinitis) and eczema (or atopic dermatitis) often coexist in the same individuals, partly because of a shared genetic origin. To identify shared risk variants, we performed a genome-wide association study (GWAS; n = 360,838) of a broad allergic disease phenotype that considers the presence of any one of these three diseases. We identified 136 independent risk variants (P < 3 × 10-8), including 73 not previously reported, which implicate 132 nearby genes in allergic disease pathophysiology. Disease-specific effects were detected for only six variants, confirming that most represent shared risk factors. Tissue-specific heritability and biological process enrichment analyses suggest that shared risk variants influence lymphocyte-mediated immunity. Six target genes provide an opportunity for drug repositioning, while for 36 genes CpG methylation was found to influence transcription independently of genetic effects. Asthma, hay fever and eczema partly coexist because they share many genetic risk variants that dysregulate the expression of immune-related genes.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
-
- Thomsen SF, et al. Findings on the atopic triad from a Danish twin registry. The international journal of tuberculosis and lung disease : the official journal of the International Union against Tuberculosis and Lung Disease. 2006;10:1268–1272. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

