Detecting positive selection in the genome
- PMID: 29084517
- PMCID: PMC5662103
- DOI: 10.1186/s12915-017-0434-y
Detecting positive selection in the genome
Abstract
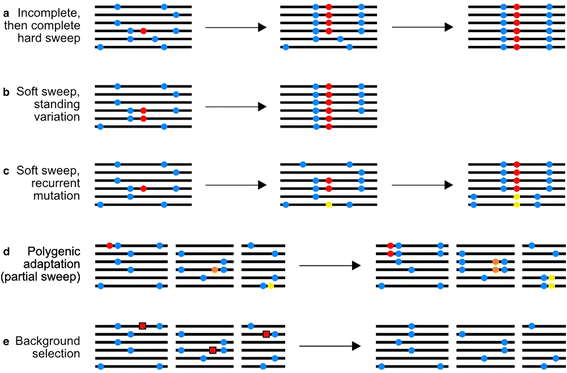
Population geneticists have long sought to understand the contribution of natural selection to molecular evolution. A variety of approaches have been proposed that use population genetics theory to quantify the rate and strength of positive selection acting in a species' genome. In this review we discuss methods that use patterns of between-species nucleotide divergence and within-species diversity to estimate positive selection parameters from population genomic data. We also discuss recently proposed methods to detect positive selection from a population's haplotype structure. The application of these tests has resulted in the detection of pervasive adaptive molecular evolution in multiple species.
Conflict of interest statement
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
-
- Kimura M. The neutral theory of molecular evolution. Cambridge University Press; 1983.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

