The multidimensional mechanisms of long noncoding RNA function
- PMID: 29084573
- PMCID: PMC5663108
- DOI: 10.1186/s13059-017-1348-2
The multidimensional mechanisms of long noncoding RNA function
Abstract
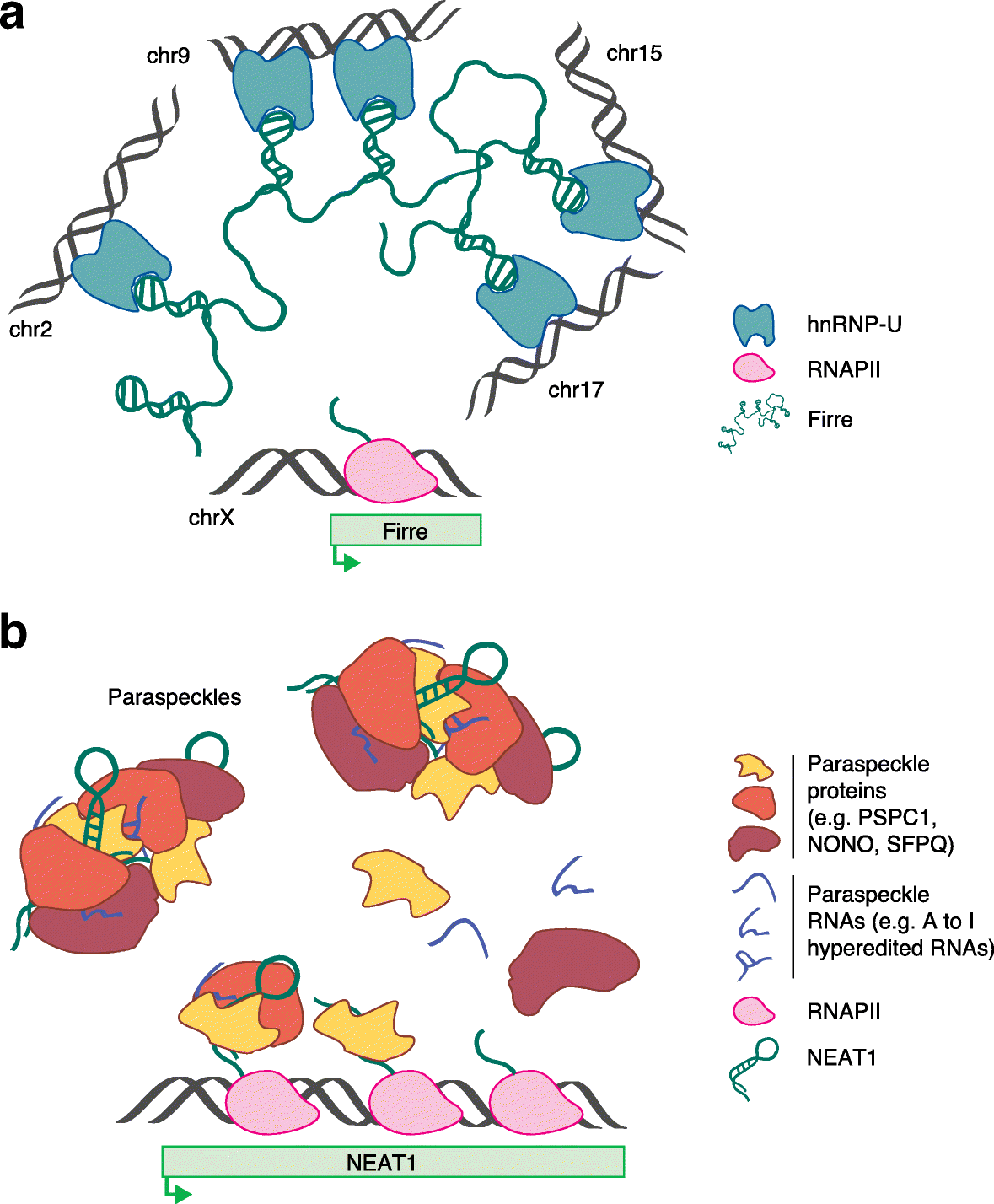
A major shift in our understanding of genome regulation has emerged recently. It is now apparent that the majority of cellular transcripts do not code for proteins, and many of them are long noncoding RNAs (lncRNAs). Increasingly, studies suggest that lncRNAs regulate gene expression through diverse mechanisms. We review emerging mechanistic views of lncRNAs in gene regulation in the cell nucleus. We discuss the functional interactions that lncRNAs establish with other molecules as well as the relationship between lncRNA transcription and function. While some of these mechanisms are specific to lncRNAs, others might be shared with other types of genes.
Conflict of interest statement
Competing interests
The authors declare that they have no competing interests.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
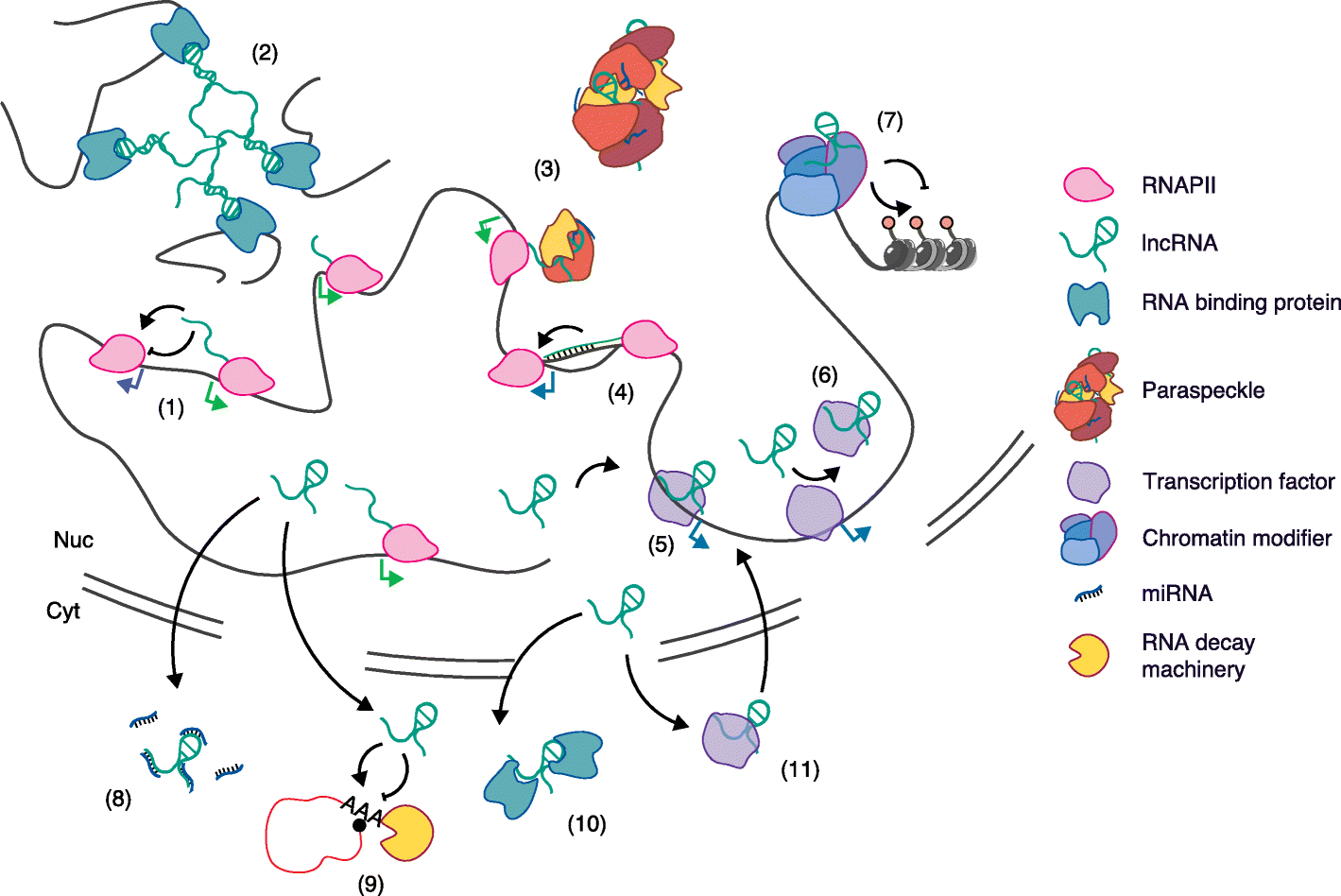
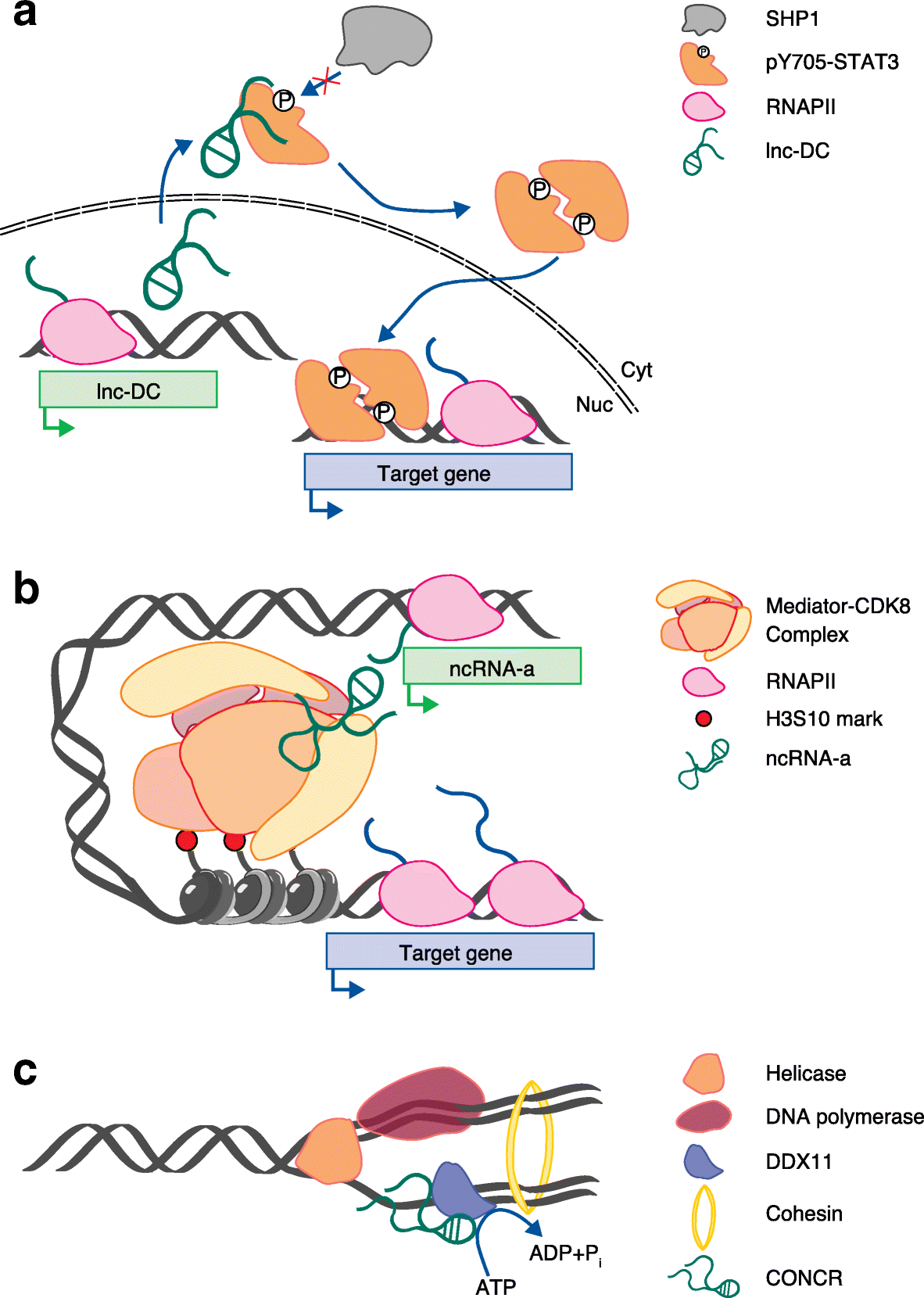
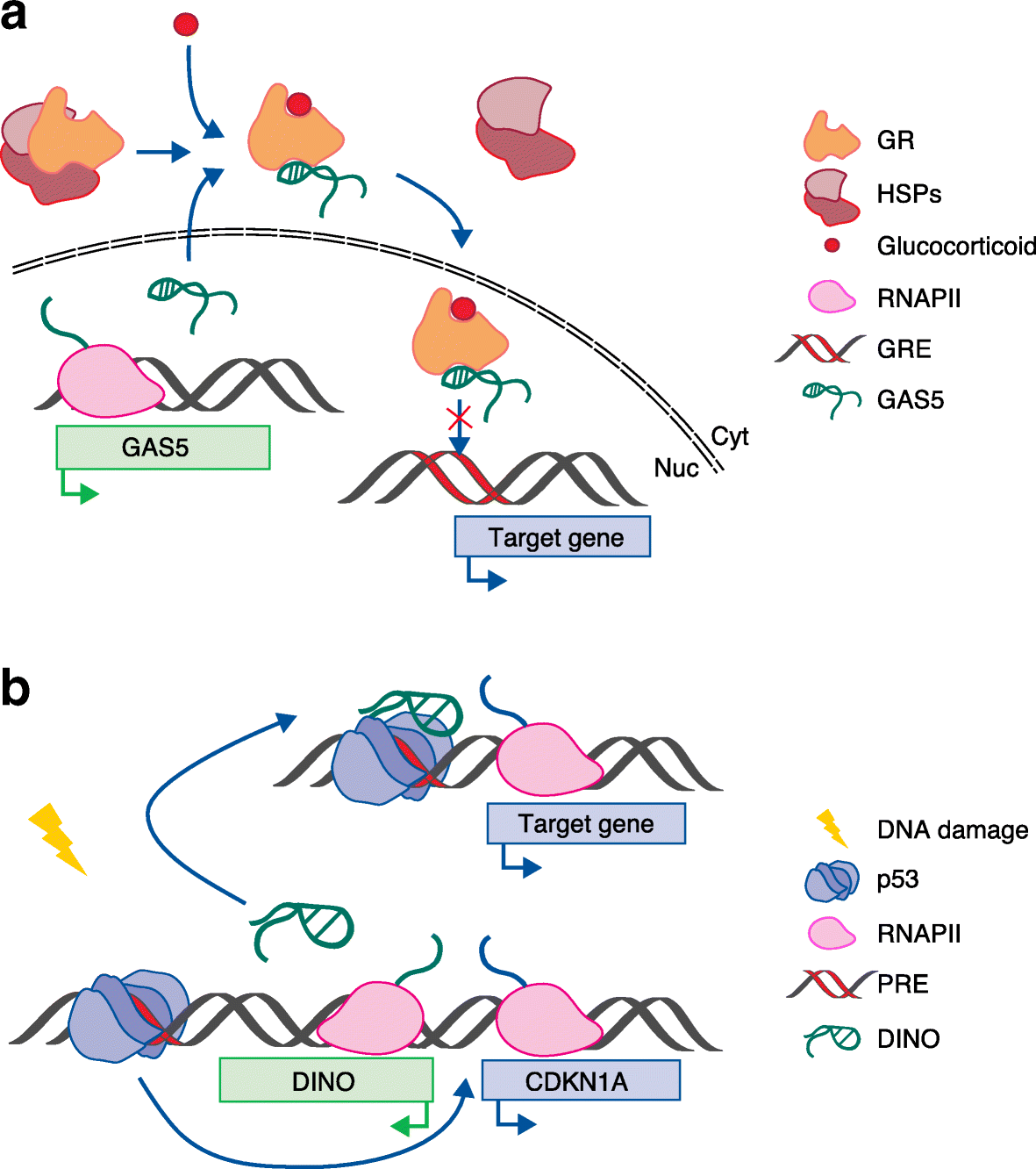
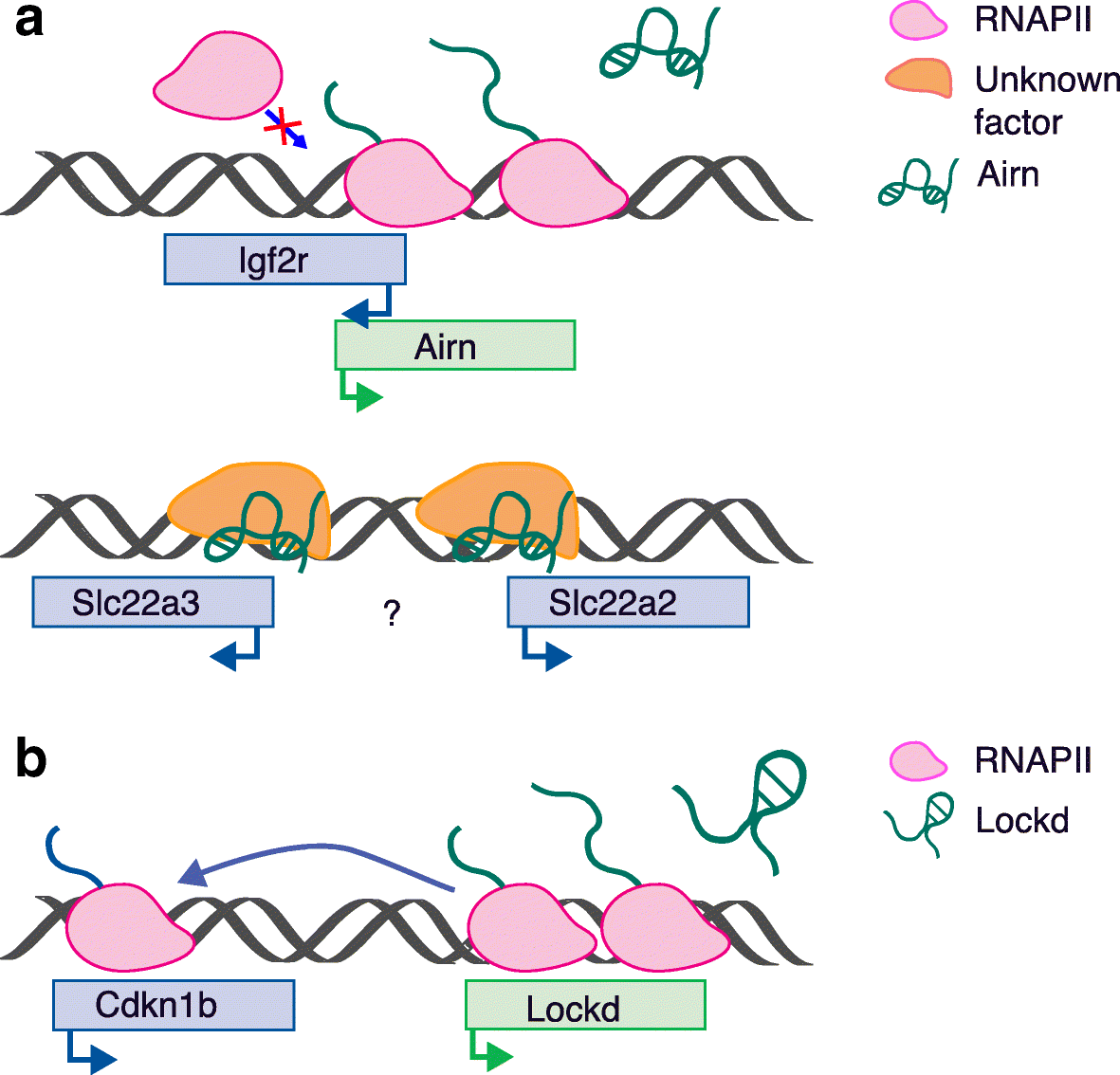
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

