Conservation and variation in pair-rule gene expression and function in the intermediate-germ beetle Dermestes maculatus
- PMID: 29084804
- PMCID: PMC5769621
- DOI: 10.1242/dev.154039
Conservation and variation in pair-rule gene expression and function in the intermediate-germ beetle Dermestes maculatus
Abstract
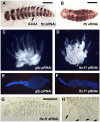
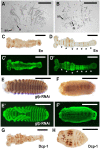
A set of pair-rule (PR) segmentation genes (PRGs) promotes the formation of alternate body segments in Drosophila melanogaster Whereas Drosophila embryos are long-germ, with segments specified more or less simultaneously, most insects add segments sequentially as the germband elongates. The hide beetle Dermestes maculatus represents an intermediate between short- and long-germ development, ideal for comparative study of PRGs. We show that eight of nine Drosophila PRG orthologs are expressed in stripes in Dermestes Functional results parse these genes into three groups: Dmac-eve, -odd and -run play roles in both germband elongation and PR patterning; Dmac-slp and -prd function exclusively as complementary, classic PRGs, supporting functional decoupling of elongation and segment formation; and orthologs of ftz, ftz-f1, h and opa show more variable function in Dermestes and other species. While extensive cell death generally prefigured Dermestes PRG RNAi-mediated cuticle defects, an organized region with high mitotic activity near the margin of the segment addition zone is likely to have contributed to truncation of eveRNAi embryos. Our results suggest general conservation of clock-like regulation of PR stripe addition in sequentially segmenting species while highlighting regulatory rewiring involving a subset of PRG orthologs.
Keywords: Beetle; Dermestes; Evo-devo; Germband elongation; Pair-rule gene; Segmentation.
© 2017. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures





References
-
- Akam M. (1987). The molecular basis for metameric pattern in the Drosophila embryo. Development 101, 1-22. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

