Substrate Specificity for Bacterial RNases HII and HIII Is Influenced by Metal Availability
- PMID: 29084857
- PMCID: PMC5786700
- DOI: 10.1128/JB.00401-17
Substrate Specificity for Bacterial RNases HII and HIII Is Influenced by Metal Availability
Abstract
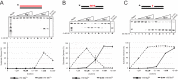
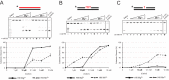
We tested the activities of four predicated RNase H enzymes, including two RNase HI-type enzymes, in addition to RNase HII (RnhB) and RNase HIII (RnhC), on several RNA-DNA hybrid substrates with different divalent metal cations. We found that the two RNase HI-type enzymes, YpdQ and YpeP, failed to show activity on the three substrates tested. RNase HII and RNase HIII cleaved all the substrates tested, although the activity was dependent on the metal made available. We show that Bacillus subtilis RNase HII and RNase HIII are both able to incise 5' to a single ribonucleoside monophosphate (rNMP). We show that RNase HIII incision at a single rNMP occurs most efficiently with Mn2+, an activity we found to be conserved among other Gram-positive RNase HIII enzymes. Characterization of RNases HII and HIII with metal concentrations in the physiological range showed that RNase HII can cleave at single rNMPs embedded in DNA while RNase HIII is far less effective. Further, using metal concentrations within the physiological range, RNase HIII efficiently cleaved longer RNA-DNA hybrids lacking an RNA-DNA junction, while RNase HII was much less effective. Phenotypic analysis showed that cells with an rnhC deletion were sensitive to hydroxyurea (HU). In contrast, cells with an rnhB deletion showed wild-type growth in the presence of HU, supporting the hypothesis that RNases HII and HIII have distinct substrate specificities in vivo This work demonstrates how metal availability influences the substrate recognition and activity of RNases HII and HIII, providing insight into their functions in vivoIMPORTANCE RNase H represents a class of proteins that cleave RNA-DNA hybrids, helping resolve R-loops and Okazaki fragments, as well as initiating the process of ribonucleotide excision repair (RER). We investigated the activities of four Bacillus subtilis RNase H enzymes and found that only RNases HII and HIII have activity and that their substrate preference is dependent on metal availability. To understand the factors that contribute to RNase HII and RNase HIII substrate preference, we show that in the presence of metal concentrations within the physiological range, RNases HII and HIII have distinct activities on different RNA-DNA hybrids. This work provides insight into how RNases HII and HIII repair the broad range of RNA-DNA hybrids that form in Gram-positive bacteria.
Keywords: Bacillus subtilis; DNA repair; R-loop; RNA-DNA hybrid; RNase; ribonucleotide excision repair.
Copyright © 2018 American Society for Microbiology.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

