Multi-dimensional genomic analysis of myoepithelial carcinoma identifies prevalent oncogenic gene fusions
- PMID: 29084941
- PMCID: PMC5662567
- DOI: 10.1038/s41467-017-01178-z
Multi-dimensional genomic analysis of myoepithelial carcinoma identifies prevalent oncogenic gene fusions
Abstract
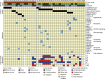
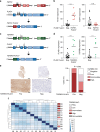
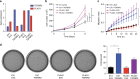
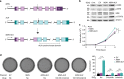
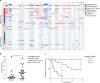
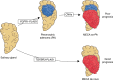
Myoepithelial carcinoma (MECA) is an aggressive salivary gland cancer with largely unknown genetic features. Here we comprehensively analyze molecular alterations in 40 MECAs using integrated genomic analyses. We identify a low mutational load, and high prevalence (70%) of oncogenic gene fusions. Most fusions involve the PLAG1 oncogene, which is associated with PLAG1 overexpression. We find FGFR1-PLAG1 in seven (18%) cases, and the novel TGFBR3-PLAG1 fusion in six (15%) cases. TGFBR3-PLAG1 promotes a tumorigenic phenotype in vitro, and is absent in 723 other salivary gland tumors. Other novel PLAG1 fusions include ND4-PLAG1; a fusion between mitochondrial and nuclear DNA. We also identify higher number of copy number alterations as a risk factor for recurrence, independent of tumor stage at diagnosis. Our findings indicate that MECA is a fusion-driven disease, nominate TGFBR3-PLAG1 as a hallmark of MECA, and provide a framework for future diagnostic and therapeutic research in this lethal cancer.
Conflict of interest statement
T.A.C. is a cofounder of Gritstone Oncology. The remaining authors declare no competing financial interests.
Figures

References
-
- Eveson J. W. A. P., Gnepp D. R., El-Naggar A. K. Pathology and Genetics of Head and Neck Tumours (International Agency for Research on Cancer, World Health Organization, Lyon, France, 2005).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

