Co-Culture of Plant Beneficial Microbes as Source of Bioactive Metabolites
- PMID: 29085019
- PMCID: PMC5662714
- DOI: 10.1038/s41598-017-14569-5
Co-Culture of Plant Beneficial Microbes as Source of Bioactive Metabolites
Abstract
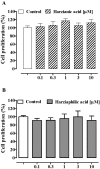
In microbial cultures the production of secondary metabolites is affected by experimental conditions, and the discovery of novel compounds is often prevented by the re-isolation of known metabolites. To limit this, it is possible to cultivate microorganisms by simulating naturally occurring interactions, where microbes co-exist in complex communities. In this work, co-culturing experiments of the biocontrol agent Trichoderma harzianum M10 and the endophyte Talaromyces pinophilus F36CF have been performed to elicit the expression of genes which are not transcribed in standard laboratory assays. Metabolomic analysis revealed that the co-culture induced the accumulation of siderophores for both fungi, while production of M10 harzianic and iso-harzianic acids was not affected by F36CF. Conversely, metabolites of the latter strain, 3-O-methylfunicone and herquline B, were less abundant when M10 was present. A novel compound, hereby named harziaphilic acid, was isolated from fungal co-cultures, and fully characterized. Moreover, harzianic and harziaphilic acids did not affect viability of colorectal cancer and healthy colonic epithelial cells, but selectively reduced cancer cell proliferation. Our results demonstrated that the co-cultivation of plant beneficial fungi may represent an effective strategy to modulate the production of bioactive metabolites and possibly identify novel compounds.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

