A cell based, high throughput assay for quantitative analysis of Hedgehog pathway activation using a Smoothened activation sensor
- PMID: 29085027
- PMCID: PMC5662767
- DOI: 10.1038/s41598-017-14767-1
A cell based, high throughput assay for quantitative analysis of Hedgehog pathway activation using a Smoothened activation sensor
Abstract
The Hedgehog (Hh) signalling cascade plays an important role in development and disease. In the absence of Hh ligand, activity of the key signal transducer Smoothened (Smo) is downregulated by the Hh receptor Patched (Ptc). However, the mechanisms underlying this inhibition, and especially its release upon ligand stimulation, are still poorly understood, in part because tools for following Smo activation at the subcellular level were long lacking. To address this deficit we have developed a high throughput cell culture assay based on a fluorescent sensor for Drosophila Smo activation. We have screened a small molecule inhibitor library, and observed increased Smo sensor fluorescence with compounds aimed at two major target groups, the MAPK signalling cascade and polo and aurora kinases. Biochemical validation for selected inhibitors (dobrafenib, tak-733, volasertib) confirmed the screen results and revealed differences in the mode of Smo activation. Furthermore, monitoring Smo activation at the single cell level indicated that individual cells exhibit different threshold responses to Hh stimulation, which may be mechanistically relevant for the formation of graded Hh responses. Together, these results thus provide proof of principle that our assay may become a valuable tool for dissecting the cell biological basis of Hh pathway activation.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
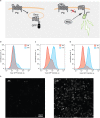
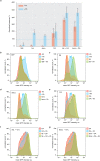

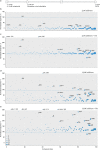
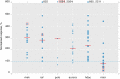

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

