Gene Acquisition by a Distinct Phyletic Group within Streptococcus pneumoniae Promotes Adhesion to the Ocular Epithelium
- PMID: 29085912
- PMCID: PMC5656748
- DOI: 10.1128/mSphere.00213-17
Gene Acquisition by a Distinct Phyletic Group within Streptococcus pneumoniae Promotes Adhesion to the Ocular Epithelium
Abstract
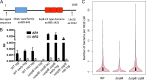
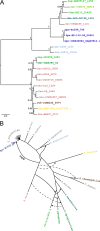
Streptococcus pneumoniae (pneumococcus) displays broad tissue tropism and infects multiple body sites in the human host. However, infections of the conjunctiva are limited to strains within a distinct phyletic group with multilocus sequence types ST448, ST344, ST1186, ST1270, and ST2315. In this study, we sequenced the genomes of six pneumococcal strains isolated from eye infections. The conjunctivitis isolates are grouped in a distinct phyletic group together with a subset of nasopharyngeal isolates. The keratitis (infection of the cornea) and endophthalmitis (infection of the vitreous body) isolates are grouped with the remainder of pneumococcal strains. Phenotypic characterization is consistent with morphological differences associated with the distinct phyletic group. Specifically, isolates from the distinct phyletic group form aggregates in planktonic cultures and chain-like structures in biofilms grown on abiotic surfaces. To begin to investigate the association between genotype and epidemiology, we focused on a predicted surface-exposed adhesin (SspB) encoded exclusively by this distinct phyletic group. Phylogenetic analysis of the gene encoding SspB in the context of a streptococcal species tree suggests that sspB was acquired by lateral gene transfer from Streptococcus suis. Furthermore, an sspB deletion mutant displays decreased adherence to cultured cells from the ocular epithelium compared to the isogenic wild-type and complemented strains. Together these findings suggest that acquisition of genes from outside the species has contributed to pneumococcal tissue tropism by enhancing the ability of a subset of strains to infect the ocular epithelium causing conjunctivitis. IMPORTANCE Changes in the gene content of pathogens can modify their ability to colonize and/or survive in different body sites in the human host. In this study, we investigate a gene acquisition event and its role in the pathogenesis of Streptococccus pneumoniae (pneumococcus). Our findings suggest that the gene encoding the predicted surface protein SspB has been transferred from Streptococcus suis (a distantly related streptococcal species) into a distinct set of pneumococcal strains. This group of strains distinguishes itself from the remainder of pneumococcal strains by extensive differences in genomic composition and by the ability to cause conjunctivitis. We find that the presence of sspB increases adherence of pneumococcus to the ocular epithelium. Thus, our data support the hypothesis that a subset of pneumococcal strains has gained genes from neighboring species that enhance their ability to colonize the epithelium of the eye, thus expanding into a new niche.
Keywords: Streptococcus pneumoniae; gene transfer; genomics; host-pathogen interactions; phylogenetic analysis.
Figures


References
-
- Donati C, Hiller NL, Tettelin H, Muzzi A, Croucher NJ, Angiuoli SV, Oggioni M, Dunning Hotopp JC, Hu FZ, Riley DR, Covacci A, Mitchell TJ, Bentley SD, Kilian M, Ehrlich GD, Rappuoli R, Moxon ER, Masignani V. 2010. Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species. Genome Biol 11:R107. doi: 10.1186/gb-2010-11-10-r107. - DOI - PMC - PubMed
-
- Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, DeBoy RT, Davidsen TM, Mora M, Scarselli M, Margarit y Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O’Connor KJB, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc Natl Acad Sci U S A 102:13950–13955. doi: 10.1073/pnas.0506758102. - DOI - PMC - PubMed
-
- Kulohoma BW, Cornick JE, Chaguza C, Yalcin F, Harris SR, Gray KJ, Kiran AM, Molyneux E, French N, Parkhill J, Faragher BE, Everett DB, Bentley SD, Heyderman RS. 2015. Comparative genomic analysis of meningitis- and bacteremia-causing pneumococci identifies a common core genome. Infect Immun 83:4165–4173. doi: 10.1128/IAI.00814-15. - DOI - PMC - PubMed
-
- Hiller NL, Janto B, Hogg JS, Boissy R, Yu S, Powell E, Keefe R, Ehrlich NE, Shen K, Hayes J, Barbadora K, Klimke W, Dernovoy D, Tatusova T, Parkhill J, Bentley SD, Post JC, Ehrlich GD, Hu FZ. 2007. Comparative genomic analyses of seventeen Streptococcus pneumoniae strains: insights into the pneumococcal supragenome. J Bacteriol 189:8186–8195. doi: 10.1128/JB.00690-07. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
