An epigenome-wide association study meta-analysis of educational attainment
- PMID: 29086770
- PMCID: PMC6372242
- DOI: 10.1038/mp.2017.210
An epigenome-wide association study meta-analysis of educational attainment
Abstract
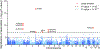
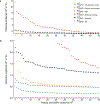
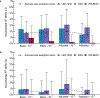
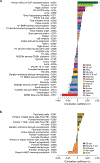
The epigenome is associated with biological factors, such as disease status, and environmental factors, such as smoking, alcohol consumption and body mass index. Although there is a widespread perception that environmental influences on the epigenome are pervasive and profound, there has been little evidence to date in humans with respect to environmental factors that are biologically distal. Here we provide evidence on the associations between epigenetic modifications-in our case, CpG methylation-and educational attainment (EA), a biologically distal environmental factor that is arguably among the most important life-shaping experiences for individuals. Specifically, we report the results of an epigenome-wide association study meta-analysis of EA based on data from 27 cohort studies with a total of 10 767 individuals. We find nine CpG probes significantly associated with EA. However, robustness analyses show that all nine probes have previously been found to be associated with smoking. Only two associations remain when we perform a sensitivity analysis in the subset of never-smokers, and these two probes are known to be strongly associated with maternal smoking during pregnancy, and thus their association with EA could be due to correlation between EA and maternal smoking. Moreover, the effect sizes of the associations with EA are far smaller than the known associations with the biologically proximal environmental factors alcohol consumption, body mass index, smoking and maternal smoking during pregnancy. Follow-up analyses that combine the effects of many probes also point to small methylation associations with EA that are highly correlated with the combined effects of smoking. If our findings regarding EA can be generalized to other biologically distal environmental factors, then they cast doubt on the hypothesis that such factors have large effects on the epigenome.
Figures


References
-
- Champagne FA, Curley JP. Epigenetic Influence of the Social Environment. In Petronis A, Mill J (ed). Brain, Behavior, and Epigenetics Springer-Verlag Berlin Heidelberg: Heidelberg, DE, 2011, pp 23–40.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

