TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions
- PMID: 29087512
- PMCID: PMC5753191
- DOI: 10.1093/nar/gkx1013
TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions
Abstract
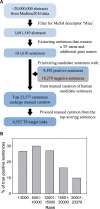
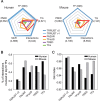
Transcription factors (TFs) are major trans-acting factors in transcriptional regulation. Therefore, elucidating TF-target interactions is a key step toward understanding the regulatory circuitry underlying complex traits such as human diseases. We previously published a reference TF-target interaction database for humans-TRRUST (Transcriptional Regulatory Relationships Unraveled by Sentence-based Text mining)-which was constructed using sentence-based text mining, followed by manual curation. Here, we present TRRUST v2 (www.grnpedia.org/trrust) with a significant improvement from the previous version, including a significantly increased size of the database consisting of 8444 regulatory interactions for 800 TFs in humans. More importantly, TRRUST v2 also contains a database for TF-target interactions in mice, including 6552 TF-target interactions for 828 mouse TFs. TRRUST v2 is also substantially more comprehensive and less biased than other TF-target interaction databases. We also improved the web interface, which now enables prioritization of key TFs for a physiological condition depicted by a set of user-input transcriptional responsive genes. With the significant expansion in the database size and inclusion of the new web tool for TF prioritization, we believe that TRRUST v2 will be a versatile database for the study of the transcriptional regulation involved in human diseases.
© The Author(s) 2017. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

