Efficient gene knockin in axolotl and its use to test the role of satellite cells in limb regeneration
- PMID: 29087939
- PMCID: PMC5703281
- DOI: 10.1073/pnas.1706855114
Efficient gene knockin in axolotl and its use to test the role of satellite cells in limb regeneration
Abstract
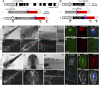
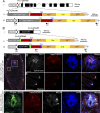
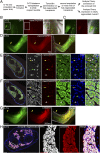
Salamanders exhibit extensive regenerative capacities and serve as a unique model in regeneration research. However, due to the lack of targeted gene knockin approaches, it has been difficult to label and manipulate some of the cell populations that are crucial for understanding the mechanisms underlying regeneration. Here we have established highly efficient gene knockin approaches in the axolotl (Ambystoma mexicanum) based on the CRISPR/Cas9 technology. Using a homology-independent method, we successfully inserted both the Cherry reporter gene and a larger membrane-tagged Cherry-ERT2-Cre-ERT2 (∼5-kb) cassette into axolotl Sox2 and Pax7 genomic loci. Depending on the size of the DNA fragments for integration, 5-15% of the F0 transgenic axolotl are positive for the transgene. Using these techniques, we have labeled and traced the PAX7-positive satellite cells as a major source contributing to myogenesis during axolotl limb regeneration. Our work brings a key genetic tool to molecular and cellular studies of axolotl regeneration.
Keywords: CRISPR/Cas9; knockin; neural stem cells; regeneration; satellite cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Kragl M, et al. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–65. - PubMed
-
- Sandoval-Guzmán T, et al. Fundamental differences in dedifferentiation and stem cell recruitment during skeletal muscle regeneration in two salamander species. Cell Stem Cell. 2014;14:174–187. - PubMed
-
- Shi Z, et al. Heritable CRISPR/Cas9-mediated targeted integration in Xenopus tropicalis. FASEB J. 2015;29:4914–4923. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

