The influence of microRNAs and poly(A) tail length on endogenous mRNA-protein complexes
- PMID: 29089021
- PMCID: PMC5664449
- DOI: 10.1186/s13059-017-1330-z
The influence of microRNAs and poly(A) tail length on endogenous mRNA-protein complexes
Abstract
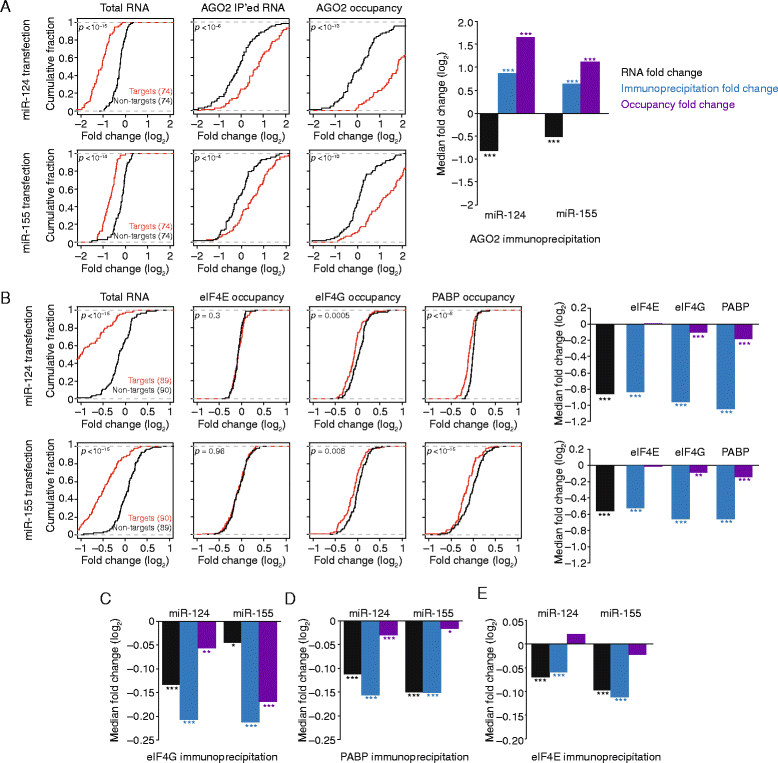
Background: All mRNAs are bound in vivo by proteins to form mRNA-protein complexes (mRNPs), but changes in the composition of mRNPs during posttranscriptional regulation remain largely unexplored. Here, we have analyzed, on a transcriptome-wide scale, how microRNA-mediated repression modulates the associations of the core mRNP components eIF4E, eIF4G, and PABP and of the decay factor DDX6 in human cells.
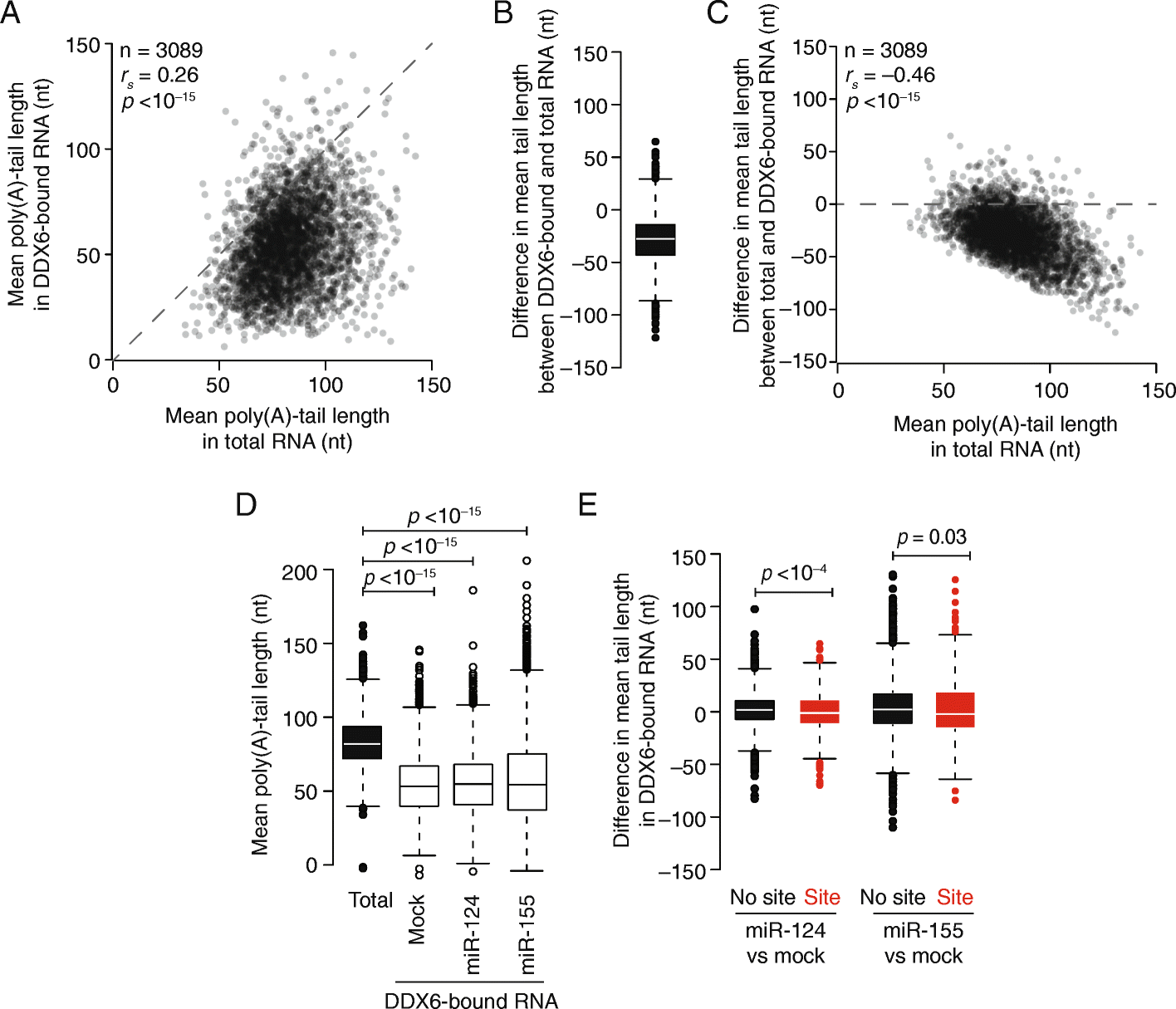
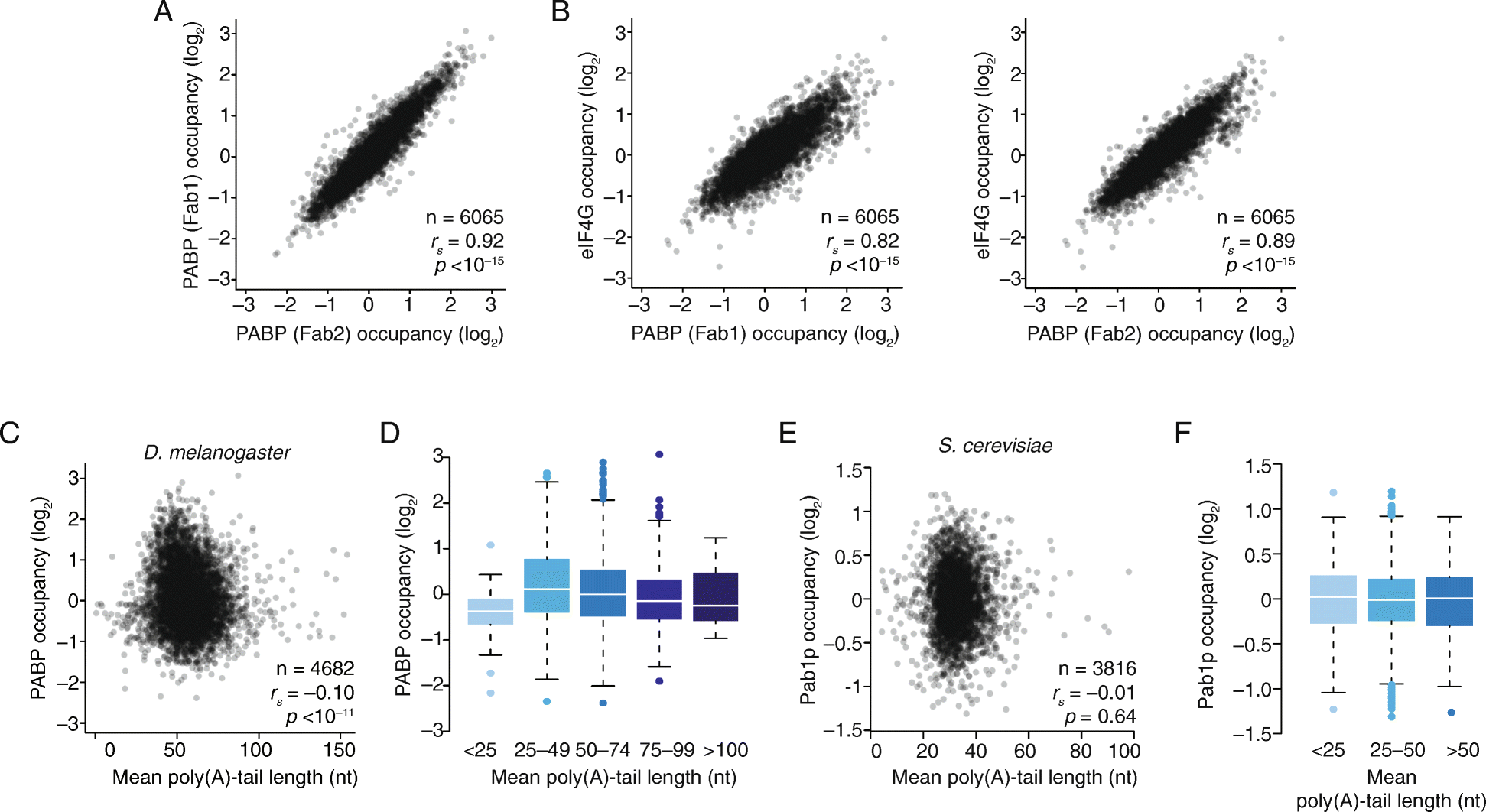
Results: Despite the transient nature of repressed intermediates, we detect significant changes in mRNP composition, marked by dissociation of eIF4G and PABP, and by recruitment of DDX6. Furthermore, although poly(A)-tail length has been considered critical in post-transcriptional regulation, differences in steady-state tail length explain little of the variation in either PABP association or mRNP organization more generally. Instead, relative occupancy of core components correlates best with gene expression.
Conclusions: These results indicate that posttranscriptional regulatory factors, such as microRNAs, influence the associations of PABP and other core factors, and do so without substantially affecting steady-state tail length.
Keywords: MicroRNAs; Poly(A) tail; mRNA–protein complexes.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

