Actionable gene-based classification toward precision medicine in gastric cancer
- PMID: 29089060
- PMCID: PMC5664811
- DOI: 10.1186/s13073-017-0484-3
Actionable gene-based classification toward precision medicine in gastric cancer
Abstract
Background: Intertumoral heterogeneity represents a significant hurdle to identifying optimized targeted therapies in gastric cancer (GC). To realize precision medicine for GC patients, an actionable gene alteration-based molecular classification that directly associates GCs with targeted therapies is needed.
Methods: A total of 207 Japanese patients with GC were included in this study. Formalin-fixed, paraffin-embedded (FFPE) tumor tissues were obtained from surgical or biopsy specimens and were subjected to DNA extraction. We generated comprehensive genomic profiling data using a 435-gene panel including 69 actionable genes paired with US Food and Drug Administration-approved targeted therapies, and the evaluation of Epstein-Barr virus (EBV) infection and microsatellite instability (MSI) status.
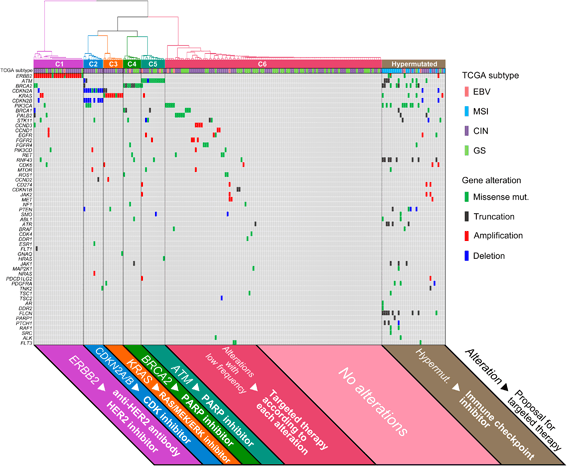
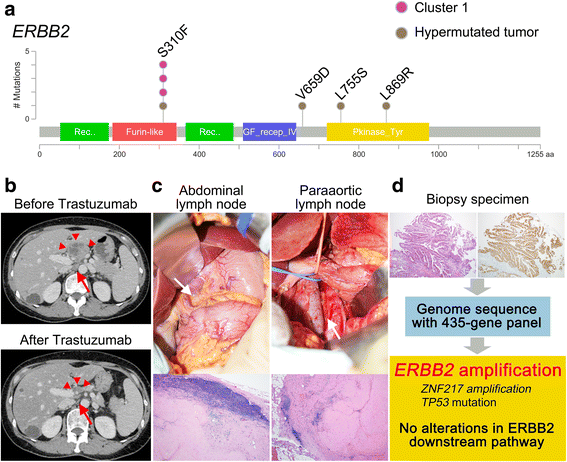
Results: Comprehensive genomic sequencing detected at least one alteration of 435 cancer-related genes in 194 GCs (93.7%) and of 69 actionable genes in 141 GCs (68.1%). We classified the 207 GCs into four The Cancer Genome Atlas (TCGA) subtypes using the genomic profiling data; EBV (N = 9), MSI (N = 17), chromosomal instability (N = 119), and genomically stable subtype (N = 62). Actionable gene alterations were not specific and were widely observed throughout all TCGA subtypes. To discover a novel classification which more precisely selects candidates for targeted therapies, 207 GCs were classified using hypermutated phenotype and the mutation profile of 69 actionable genes. We identified a hypermutated group (N = 32), while the others (N = 175) were sub-divided into six clusters including five with actionable gene alterations: ERBB2 (N = 25), CDKN2A, and CDKN2B (N = 10), KRAS (N = 10), BRCA2 (N = 9), and ATM cluster (N = 12). The clinical utility of this classification was demonstrated by a case of unresectable GC with a remarkable response to anti-HER2 therapy in the ERBB2 cluster.
Conclusions: This actionable gene-based classification creates a framework for further studies for realizing precision medicine in GC.
Keywords: Actionable gene; Gastric cancer; Gene panel; Next-generation sequencing; Precision medicine.
Conflict of interest statement
Ethics approval and consent to participate
This study was conducted in accordance with the provisions of the Declaration of Helsinki. Collection and use of all specimens in this study were approved by the Institutional Review Boards of Niigata University Graduate School of Medical and Dental Sciences (#771), Niigata Cancer Center Hospital (#641), Gifu University (#27-326), Kyushu University (#672-00), and Keio University (#20150469). Informed consent was obtained from all participants.
Consent for publication
Informed consent for the presentation of case reports was obtained from patients.
Competing interests
SL and DV are employees of and have been granted stock options by KEW Inc. The remaining authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
-
- Fujitani K, Yang HK, Mizusawa J, Kim YW, Terashima M, Han SU, et al. Gastrectomy plus chemotherapy versus chemotherapy alone for advanced gastric cancer with a single non-curable factor (REGATTA): a phase 3, randomised controlled trial. Lancet Oncol. 2016;17:309–18. doi: 10.1016/S1470-2045(15)00553-7. - DOI - PubMed
-
- Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–97. doi: 10.1016/S0140-6736(10)61121-X. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

