The Unsolved Problem of How Cells Sense Micron-Scale Curvature
- PMID: 29089160
- PMCID: PMC5705049
- DOI: 10.1016/j.tibs.2017.10.001
The Unsolved Problem of How Cells Sense Micron-Scale Curvature
Abstract
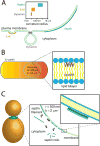
Membrane curvature is a fundamental feature of cells and their organelles. Much of what we know about how cells sense curved surfaces comes from studies examining nanometer-sized molecules on nanometer-scale curvatures. We are only just beginning to understand how cells recognize curved topologies at the micron scale. In this review, we provide the reader with an overview of our current understanding of how cells sense and respond to micron-scale membrane curvature.
Keywords: BAR domain; SpoVM; membrane curvature; septin.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

