Transcriptomes of major renal collecting duct cell types in mouse identified by single-cell RNA-seq
- PMID: 29089413
- PMCID: PMC5699061
- DOI: 10.1073/pnas.1710964114
Transcriptomes of major renal collecting duct cell types in mouse identified by single-cell RNA-seq
Abstract
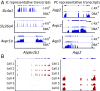
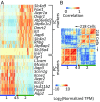
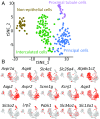
Prior RNA sequencing (RNA-seq) studies have identified complete transcriptomes for most renal epithelial cell types. The exceptions are the cell types that make up the renal collecting duct, namely intercalated cells (ICs) and principal cells (PCs), which account for only a small fraction of the kidney mass, but play critical physiological roles in the regulation of blood pressure, extracellular fluid volume, and extracellular fluid composition. To enrich these cell types, we used FACS that employed well-established lectin cell surface markers for PCs and type B ICs, as well as a newly identified cell surface marker for type A ICs, c-Kit. Single-cell RNA-seq using the IC- and PC-enriched populations as input enabled identification of complete transcriptomes of A-ICs, B-ICs, and PCs. The data were used to create a freely accessible online gene-expression database for collecting duct cells. This database allowed identification of genes that are selectively expressed in each cell type, including cell-surface receptors, transcription factors, transporters, and secreted proteins. The analysis also identified a small fraction of hybrid cells expressing aquaporin-2 and anion exchanger 1 or pendrin transcripts. In many cases, mRNAs for receptors and their ligands were identified in different cells (e.g., Notch2 chiefly in PCs vs. Jag1 chiefly in ICs), suggesting signaling cross-talk among the three cell types. The identified patterns of gene expression among the three types of collecting duct cells provide a foundation for understanding physiological regulation and pathophysiology in the renal collecting duct.
Keywords: intercalated cell; kidney; principal cell; systems biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Comment in
-
Gene expression: Single-cell RNA sequencing of collecting duct cells.Nat Rev Nephrol. 2018 Jan;14(1):3. doi: 10.1038/nrneph.2017.163. Epub 2017 Nov 20. Nat Rev Nephrol. 2018. PMID: 29151598 No abstract available.
-
Reply to Edemir: Physiological regulation and single-cell RNA sequencing.Proc Natl Acad Sci U S A. 2018 Jan 16;115(3):E351-E352. doi: 10.1073/pnas.1720333115. Epub 2018 Jan 8. Proc Natl Acad Sci U S A. 2018. PMID: 29311339 Free PMC article. No abstract available.
-
Considering hypertonicity in the interpretation and analysis of cell type-specific gene expression pattern in the collecting duct.Proc Natl Acad Sci U S A. 2018 Jan 16;115(3):E349-E350. doi: 10.1073/pnas.1720087115. Epub 2018 Jan 8. Proc Natl Acad Sci U S A. 2018. PMID: 29311340 Free PMC article. No abstract available.
References
-
- Kriz W, Bankir L. The Renal Commission of the International Union of Physiological Sciences (IUPS) A standard nomenclature for structures of the kidney. Kidney Int. 1988;33:1–7. - PubMed
-
- Cheval L, et al. Atlas of gene expression in the mouse kidney: New features of glomerular parietal cells. Physiol Genomics. 2011;43:161–173. - PubMed
-
- Zeisel A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

