Identifying host regulators and inhibitors of liver stage malaria infection using kinase activity profiles
- PMID: 29089541
- PMCID: PMC5663700
- DOI: 10.1038/s41467-017-01345-2
Identifying host regulators and inhibitors of liver stage malaria infection using kinase activity profiles
Abstract
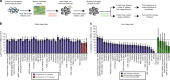
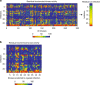
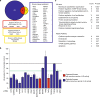
Plasmodium parasites have extensive needs from their host hepatocytes during the obligate liver stage of infection, yet there remains sparse knowledge of specific host regulators. Here we assess 34 host-targeted kinase inhibitors for their capacity to eliminate Plasmodium yoelii-infected hepatocytes. Using pre-existing activity profiles of each inhibitor, we generate a predictive computational model that identifies host kinases, which facilitate Plasmodium yoelii liver stage infection. We predict 47 kinases, including novel and previously described kinases that impact infection. The impact of a subset of kinases is experimentally validated, including Receptor Tyrosine Kinases, members of the MAP Kinase cascade, and WEE1. Our approach also predicts host-targeted kinase inhibitors of infection, including compounds already used in humans. Three of these compounds, VX-680, Roscovitine and Sunitinib, each eliminate >85% of infection. Our approach is well-suited to uncover key host determinants of infection in difficult model systems, including field-isolated parasites and/or emerging pathogens.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

