A total transcriptome profiling method for plasma-derived extracellular vesicles: applications for liquid biopsies
- PMID: 29089558
- PMCID: PMC5663969
- DOI: 10.1038/s41598-017-14264-5
A total transcriptome profiling method for plasma-derived extracellular vesicles: applications for liquid biopsies
Abstract
Extracellular vesicles (EVs) are key mediators of intercellular communication. Part of their biological effects can be attributed to the transfer of cargos of diverse types of RNAs, which are promising diagnostic and prognostic biomarkers. EVs found in human biofluids are a valuable source for the development of minimally invasive assays. However, the total transcriptional landscape of EVs is still largely unknown. Here we develop a new method for total transcriptome profiling of plasma-derived EVs by next generation sequencing (NGS) from limited quantities of patient-derived clinical samples, which enables the unbiased characterization of the complete RNA cargo, including both small- and long-RNAs, in a single library preparation step. This approach was applied to RNA extracted from EVs isolated by ultracentrifugation from the plasma of five healthy volunteers. Among the most abundant RNAs identified we found small RNAs such as tRNAs, miRNAs and miscellaneous RNAs, which have largely unknown functions. We also identified protein-coding and long noncoding transcripts, as well as circular RNA species that were also experimentally validated. This method enables, for the first time, the full spectrum of transcriptome data to be obtained from minute patient-derived samples, and will therefore potentially allow the identification of cell-to-cell communication mechanisms and biomarkers.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
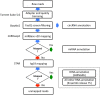


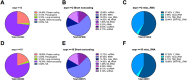
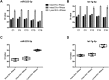
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

