High throughput discovery of influenza virus neutralizing antibodies from phage-displayed synthetic antibody libraries
- PMID: 29089574
- PMCID: PMC5663709
- DOI: 10.1038/s41598-017-14823-w
High throughput discovery of influenza virus neutralizing antibodies from phage-displayed synthetic antibody libraries
Abstract
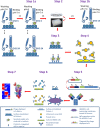
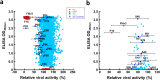
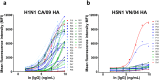
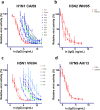
Pandemic and epidemic outbreaks of influenza A virus (IAV) infection pose severe challenges to human society. Passive immunotherapy with recombinant neutralizing antibodies can potentially mitigate the threats of IAV infection. With a high throughput neutralizing antibody discovery platform, we produced artificial anti-hemagglutinin (HA) IAV-neutralizing IgGs from phage-displayed synthetic scFv libraries without necessitating prior memory of antibody-antigen interactions or relying on affinity maturation essential for in vivo immune systems to generate highly specific neutralizing antibodies. At least two thirds of the epitope groups of the artificial anti-HA antibodies resemble those of natural protective anti-HA antibodies, providing alternatives to neutralizing antibodies from natural antibody repertoires. With continuing advancement in designing and constructing synthetic scFv libraries, this technological platform is useful in mitigating not only the threats of IAV pandemics but also those from other newly emerging viral infections.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

