Molecular Insights into Antimicrobial Resistance Traits of Multidrug Resistant Enteric Pathogens isolated from India
- PMID: 29089611
- PMCID: PMC5663842
- DOI: 10.1038/s41598-017-14791-1
Molecular Insights into Antimicrobial Resistance Traits of Multidrug Resistant Enteric Pathogens isolated from India
Abstract
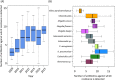
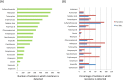
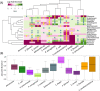
Emergence of antimicrobial resistant Gram-negative bacteria has created a serious global health crisis and threatens the effectiveness of most, if not all, antibiotics commonly used to prevent and treat bacterial infections. There is a dearth of detailed studies on the prevalence of antimicrobial resistance (AMR) patterns in India. Here, we have isolated and examined AMR patterns of 654 enteric pathogens and investigated complete genome sequences of isolates from six representative genera, which in aggregate encode resistance against 22 antibiotics representing nine distinct drug classes. This study revealed that ~97% isolates are resistant against ≥2 antibiotics, ~24% isolates are resistant against ≥10 antibiotics and ~3% isolates are resistant against ≥15 antibiotics. Analyses of whole genome sequences of six extensive drug resistant enteric pathogens revealed presence of multiple mobile genetic elements, which are physically linked with resistance traits. These elements are therefore appearing to be responsible for disseminating drug resistance among bacteria through horizontal gene transfer. The present study provides insights into the linkages between the resistance patterns to certain antibiotics and their usage in India. The findings would be useful to understand the genetics of resistance traits and severity of and difficulty in tackling AMR enteric pathogens.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- World Health Organization. Antimicrobial resistance: Global report on surveillance (2014).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

