Design, Optimization and Application of Small Molecule Biosensor in Metabolic Engineering
- PMID: 29089935
- PMCID: PMC5651080
- DOI: 10.3389/fmicb.2017.02012
Design, Optimization and Application of Small Molecule Biosensor in Metabolic Engineering
Abstract
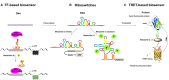
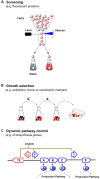
The development of synthetic biology and metabolic engineering has painted a great future for the bio-based economy, including fuels, chemicals, and drugs produced from renewable feedstocks. With the rapid advance of genome-scale modeling, pathway assembling and genome engineering/editing, our ability to design and generate microbial cell factories with various phenotype becomes almost limitless. However, our lack of ability to measure and exert precise control over metabolite concentration related phenotypes becomes a bottleneck in metabolic engineering. Genetically encoded small molecule biosensors, which provide the means to couple metabolite concentration to measurable or actionable outputs, are highly promising solutions to the bottleneck. Here we review recent advances in the design, optimization and application of small molecule biosensor in metabolic engineering, with particular focus on optimization strategies for transcription factor (TF) based biosensors.
Keywords: industrial application; metabolic engineering; optimization strategy; small molecule biosensor; synthetic biology; transcription factor.
Figures
References
-
- Ameen S., Ahmad M., Mohsin M., Qureshi M. I., Ibrahim M. M., Abdin M. Z., et al. (2016). Designing, construction and characterization of genetically encoded FRET-based nanosensor for real time monitoring of lysine flux in living cells. J. Nanobiotechnology 14:49. 10.1186/s12951-016-0204-y - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

