%MinMax: A versatile tool for calculating and comparing synonymous codon usage and its impact on protein folding
- PMID: 29090506
- PMCID: PMC5734269
- DOI: 10.1002/pro.3336
%MinMax: A versatile tool for calculating and comparing synonymous codon usage and its impact on protein folding
Abstract
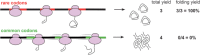
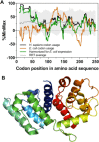
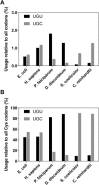
Most amino acids can be encoded by more than one synonymous codon, but these are rarely used with equal frequency. In many coding sequences the usage patterns of rare versus common synonymous codons is nonrandom and under selection. Moreover, synonymous substitutions that alter these patterns can have a substantial impact on the folding efficiency of the encoded protein. This has ignited broad interest in exploring synonymous codon usage patterns. For many protein chemists, biophysicists and structural biologists, the primary motivation for codon analysis is identifying and preserving usage patterns most likely to impact high-yield production of functional proteins. Here we describe the core functions and new features of %MinMax, a codon usage calculator freely available as a web-based portal and downloadable script (http://www.codons.org). %MinMax evaluates the relative usage frequencies of the synonymous codons used to encode a protein sequence of interest and compares these results to a rigorous null model. Crucially, for analyzing codon usage in common host organisms %MinMax requires only the coding sequence as input; with a user-input codon frequency table, %MinMax can be used to evaluate synonymous codon usage patterns for any coding sequence from any fully sequenced genome. %MinMax makes no assumptions regarding the impact of transfer ribonucleic acid concentrations or other molecular-level interactions on translation rates, yet its output is sufficient to predict the effects of synonymous codon substitutions on cotranslational folding mechanisms. A simple calculation included within %MinMax can be used to harmonize codon usage frequencies for heterologous gene expression.
Keywords: biotechnology; codon harmonization; codon optimization; cotranslational protein folding; heterologous gene expression; protein aggregation; ribosome.
© 2017 The Protein Society.
Figures
References
-
- Komar AA, Lesnik T, Reiss C (1999) Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett 462:387–391. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

