Analysis of Disulfide Bond Formation
- PMID: 29091273
- PMCID: PMC5743216
- DOI: 10.1002/cpps.43
Analysis of Disulfide Bond Formation
Abstract
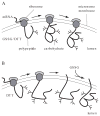
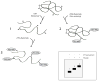
In this unit, protocols are provided for detection of disulfide bond formation in cultures of intact cells and in an in vitro translation system containing isolated microsomes or semi-permeabilized cells. First, the newly synthesized protein of interest is biosynthetically labeled with radioactive amino acids in a short pulse. The labeled protein then is chased with unlabeled amino acids. At different times during the chase, a sample is collected, membranes are lysed with detergent, and the protein is isolated by immunoprecipitation, as described. A support protocol is provided for analysis of disulfide bonds in the immunoprecipitates by SDS-PAGE with and without prior reduction. The difference in mobility observed between the gels with nonreduced and reduced samples is due to disulfide bonds in the nonreduced protein. An additional support protocol is included that uses PEG-maleimide to modify free thiols and follow disulfide-bond formation by SDS-PAGE. © 2017 by John Wiley & Sons, Inc.
Keywords: disulfide bonds; endoplasmic reticulum; protein folding; pulse-chase; radiolabeling; secretory pathway.
Copyright © 2017 John Wiley & Sons, Inc.
Figures
References
-
- Braakman I, Helenius J, Helenius A. Role of ATP and disulphide bonds during protein folding in the endoplasmic reticulum. Nature. 1992b;356:260–262. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

