Low-Grade Astrocytoma Mutations in IDH1, P53, and ATRX Cooperate to Block Differentiation of Human Neural Stem Cells via Repression of SOX2
- PMID: 29091765
- PMCID: PMC5687844
- DOI: 10.1016/j.celrep.2017.10.009
Low-Grade Astrocytoma Mutations in IDH1, P53, and ATRX Cooperate to Block Differentiation of Human Neural Stem Cells via Repression of SOX2
Abstract
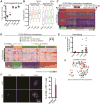
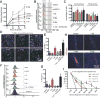
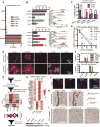
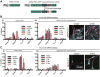
Low-grade astrocytomas (LGAs) carry neomorphic mutations in isocitrate dehydrogenase (IDH) concurrently with P53 and ATRX loss. To model LGA formation, we introduced R132H IDH1, P53 shRNA, and ATRX shRNA into human neural stem cells (NSCs). These oncogenic hits blocked NSC differentiation, increased invasiveness in vivo, and led to a DNA methylation and transcriptional profile resembling IDH1 mutant human LGAs. The differentiation block was caused by transcriptional silencing of the transcription factor SOX2 secondary to disassociation of its promoter from a putative enhancer. This occurred because of reduced binding of the chromatin organizer CTCF to its DNA motifs and disrupted chromatin looping. Our human model of IDH mutant LGA formation implicates impaired NSC differentiation because of repression of SOX2 as an early driver of gliomagenesis.
Keywords: ATRX; CTCF; DNA methylation; IDH; P53; SOX2; astrocytoma; chromatin looping; low-grade glioma; neural stem cells.
Copyright © 2017 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Amador-Arjona A, Cimadamore F, Huang CT, Wright R, Lewis S, Gage FH, Terskikh AV. SOX2 primes the epigenetic landscape in neural precursors enabling proper gene activation during hippocampal neurogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E1936–1945. - PMC - PubMed
-
- Amary MF, Damato S, Halai D, Eskandarpour M, Berisha F, Bonar F, McCarthy S, Fantin VR, Straley KS, Lobo S, et al. Ollier disease and Maffucci syndrome are caused by somatic mosaic mutations of IDH1 and IDH2. Nat Genet. 2011;43:1262–1265. - PubMed
-
- Annovazzi L, Mellai M, Caldera V, Valente G, Schiffer D. SOX2 expression and amplification in gliomas and glioma cell lines. Cancer Genomics Proteomics. 2011;8:139–147. - PubMed
-
- Baer K, Eriksson PS, Faull RL, Rees MI, Curtis MA. Sox-2 is expressed by glial and progenitor cells and Pax-6 is expressed by neuroblasts in the human subventricular zone. Exp Neurol. 2007;204:828–831. - PubMed
MeSH terms
Substances
Grants and funding
- UL1 TR001445/TR/NCATS NIH HHS/United States
- T32 CA009161/CA/NCI NIH HHS/United States
- P30 CA016087/CA/NCI NIH HHS/United States
- R01 GM112192/GM/NIGMS NIH HHS/United States
- T32 GM007308/GM/NIGMS NIH HHS/United States
- S10 OD018338/OD/NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- R21 CA188968/CA/NCI NIH HHS/United States
- R01 GM064844/GM/NIGMS NIH HHS/United States
- P30 NS050276/NS/NINDS NIH HHS/United States
- K99 GM117302/GM/NIGMS NIH HHS/United States
- R01 GM115384/GM/NIGMS NIH HHS/United States
- R21 NS088775/NS/NINDS NIH HHS/United States
- R01 GM086852/GM/NIGMS NIH HHS/United States
- R21 NS087241/NS/NINDS NIH HHS/United States
- R03 NS087349/NS/NINDS NIH HHS/United States
- UL1 TR000038/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

