Single-Cell RNA-Seq Analysis of Infiltrating Neoplastic Cells at the Migrating Front of Human Glioblastoma
- PMID: 29091775
- PMCID: PMC5810554
- DOI: 10.1016/j.celrep.2017.10.030
Single-Cell RNA-Seq Analysis of Infiltrating Neoplastic Cells at the Migrating Front of Human Glioblastoma
Abstract
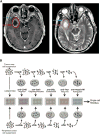
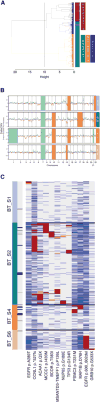
Glioblastoma (GBM) is the most common primary brain cancer in adults and is notoriously difficult to treat because of its diffuse nature. We performed single-cell RNA sequencing (RNA-seq) on 3,589 cells in a cohort of four patients. We obtained cells from the tumor core as well as surrounding peripheral tissue. Our analysis revealed cellular variation in the tumor's genome and transcriptome. We were also able to identify infiltrating neoplastic cells in regions peripheral to the core lesions. Despite the existence of significant heterogeneity among neoplastic cells, we found that infiltrating GBM cells share a consistent gene signature between patients, suggesting a common mechanism of infiltration. Additionally, in investigating the immunological response to the tumors, we found transcriptionally distinct myeloid cell populations residing in the tumor core and the surrounding peritumoral space. Our data provide a detailed dissection of GBM cell types, revealing an abundance of information about tumor formation and migration.
Keywords: GBM; RNA-seq; brain; checkpoint; diffuse; glioblastoma; glioma; heterogeneity; infiltrating; single cell.
Copyright © 2017 Elsevier Inc. All rights reserved
Figures



References
-
- Bush NAO, Chang SM, Berger MS. Current and future strategies for treatment of glioma. Neurosurg Rev. 2016;40:1–14. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

