Germline competency of human embryonic stem cells depends on eomesodermin
- PMID: 29091993
- PMCID: PMC5803789
- DOI: 10.1093/biolre/iox138
Germline competency of human embryonic stem cells depends on eomesodermin
Abstract
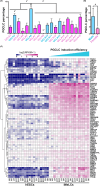
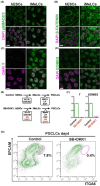
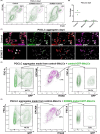
In humans, germline competency and the specification of primordial germ cells (PGCs) are thought to occur in a restricted developmental window during early embryogenesis. Despite the importance of specifying the appropriate number of PGCs for human reproduction, the molecular mechanisms governing PGC formation remain largely unexplored. Here, we compared PGC-like cell (PGCLC) differentiation from 18 independently derived human embryonic stem cell (hESC) lines, and discovered that the expression of primitive streak genes were positively associated with hESC germline competency. Furthermore, we show that chemical inhibition of TGFβ and WNT signaling, which are required for primitive streak formation and CRISPR/Cas9 deletion of Eomesodermin (EOMES), significantly impacts PGCLC differentiation from hESCs. Taken together, our results suggest that human PGC formation involves signaling and transcriptional programs associated with somatic germ layer induction and expression of EOMES.
Keywords: EOMES; embryonic stem cells; human; primordial germ cells.
© The Authors 2017. Published by Oxford University Press on behalf of Society for the Study of Reproduction. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures

References
-
- Magnusdottir E, Surani MA. How to make a primordial germ cell. Development 2014; 141:245–252. - PubMed
-
- Tang WWC, Kobayashi T, Irie N, Dietmann S, Surani MA. Specification and epigenetic programming of the human germ line. Nat Rev Genet 2016; 17:585–600. - PubMed
-
- Sasaki K, Nakamura T, Okamoto I, Yabuta Y, Iwatani C, Tsuchiya H, Seita Y, Nakamura S, Shiraki N, Takakuwa T, Yamamoto T, Saitou M. The germ cell fate of cynomolgus monkeys is specified in the nascent amnion. Dev Cell 2016; 39:169–185. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

