TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells
- PMID: 29092952
- PMCID: PMC6042652
- DOI: 10.1158/0008-5472.CAN-17-0307
TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells
Abstract
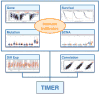
Recent clinical successes of cancer immunotherapy necessitate the investigation of the interaction between malignant cells and the host immune system. However, elucidation of complex tumor-immune interactions presents major computational and experimental challenges. Here, we present Tumor Immune Estimation Resource (TIMER; cistrome.shinyapps.io/timer) to comprehensively investigate molecular characterization of tumor-immune interactions. Levels of six tumor-infiltrating immune subsets are precalculated for 10,897 tumors from 32 cancer types. TIMER provides 6 major analytic modules that allow users to interactively explore the associations between immune infiltrates and a wide spectrum of factors, including gene expression, clinical outcomes, somatic mutations, and somatic copy number alterations. TIMER provides a user-friendly web interface for dynamic analysis and visualization of these associations, which will be of broad utilities to cancer researchers. Cancer Res; 77(21); e108-10. ©2017 AACR.
©2017 American Association for Cancer Research.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures
References
-
- Hackl H, Charoentong P, Finotello F, Trajanoski Z. Computational genomics tools for dissecting tumour-immune cell interactions. Nature reviews Genetics. 2016;17:441–58. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

