DeepPhe: A Natural Language Processing System for Extracting Cancer Phenotypes from Clinical Records
- PMID: 29092954
- PMCID: PMC5690492
- DOI: 10.1158/0008-5472.CAN-17-0615
DeepPhe: A Natural Language Processing System for Extracting Cancer Phenotypes from Clinical Records
Abstract
Precise phenotype information is needed to understand the effects of genetic and epigenetic changes on tumor behavior and responsiveness. Extraction and representation of cancer phenotypes is currently mostly performed manually, making it difficult to correlate phenotypic data to genomic data. In addition, genomic data are being produced at an increasingly faster pace, exacerbating the problem. The DeepPhe software enables automated extraction of detailed phenotype information from electronic medical records of cancer patients. The system implements advanced Natural Language Processing and knowledge engineering methods within a flexible modular architecture, and was evaluated using a manually annotated dataset of the University of Pittsburgh Medical Center breast cancer patients. The resulting platform provides critical and missing computational methods for computational phenotyping. Working in tandem with advanced analysis of high-throughput sequencing, these approaches will further accelerate the transition to precision cancer treatment. Cancer Res; 77(21); e115-8. ©2017 AACR.
©2017 American Association for Cancer Research.
Figures
References
-
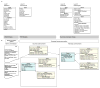
- DeepPhe Information Model [Internet] Available from: https://github.com/DeepPhe/models.
-
- UIMA [Internet] 2013 Available from: uima.apache.org.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

