Rabbit Hemorrhagic Disease Virus 2 (RHDV2; GI.2) Is Replacing Endemic Strains of RHDV in the Australian Landscape within 18 Months of Its Arrival
- PMID: 29093089
- PMCID: PMC5752944
- DOI: 10.1128/JVI.01374-17
Rabbit Hemorrhagic Disease Virus 2 (RHDV2; GI.2) Is Replacing Endemic Strains of RHDV in the Australian Landscape within 18 Months of Its Arrival
Abstract
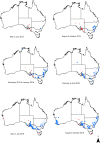
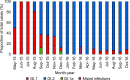
Rabbit hemorrhagic disease virus 2 (RHDV2; Lagovirus GI.2) is a pathogenic calicivirus that affects European rabbits (Oryctolagus cuniculus) and various hare (Lepus) species. GI.2 was first detected in France in 2010 and subsequently caused epidemics in wild and domestic lagomorph populations throughout Europe. In May 2015, GI.2 was detected in Australia. Within 18 months of its initial detection, GI.2 had spread to all Australian states and territories and rapidly became the dominant circulating strain, replacing Rabbit hemorrhagic disease virus (RHDV/GI.1) in mainland Australia. Reconstruction of the evolutionary history of 127 Australian GI.2 isolates revealed that the virus arrived in Australia at least several months before its initial description and likely circulated unnoticed in wild rabbit populations in the east of the continent prior to its detection. GI.2 sequences isolated from five hares clustered with sequences from sympatric rabbit populations sampled contemporaneously, indicating multiple spillover events into hares rather than an adaptation of the Australian GI.2 to a new host. Since the presence of GI.2 in Australia may have wide-ranging consequences for rabbit biocontrol, particularly with the release of the novel biocontrol agent GI.1a/RHDVa-K5 in March 2017, ongoing surveillance is critical to understanding the interactions of the various lagoviruses in Australia and their impact on host populations.IMPORTANCE This study describes the spread and distribution of Rabbit hemorrhagic disease virus 2 (GI.2) in Australia since its first detection in May 2015. Within the first 18 months following its detection, RHDV2 spread from east to west across the continent and became the dominant strain in all mainland states of Australia. This has important implications for pest animal management and for owners of pet and farmed rabbits, as there currently is no effective vaccine available in Australia for GI.2. The closely related RHDV (GI.1) is used to control overabundant wild rabbits, a serious environmental and agricultural pest in this country, and it is currently unclear how the widespread circulation of GI.2 will impact ongoing targeted wild rabbit management operations.
Keywords: RHDV2; biocontrol; calicivirus; distribution; establishment; evolution; rabbit hemorrhagic disease virus.
Copyright © 2018 American Society for Microbiology.
Figures



References
-
- Camarda A, Pugliese N, Cavadini P, Circella E, Capucci L, Caroli A, Legretto M, Mallia E, Lavazza A. 2014. Detection of the new emerging rabbit haemorrhagic disease type 2 virus (RHDV2) in Sicily from rabbit (Oryctolagus cuniculus) and Italian hare (Lepus corsicanus). Res Vet Sci 97:642–645. doi: 10.1016/j.rvsc.2014.10.008. - DOI - PubMed
-
- Le Pendu J, Abrantes J, Bertagnoli S, Guitton JS, Le Gall-Recule G, Lopes AM, Marchandeau S, Alda F, Almeida T, Celio AP, Barcena J, Burmakina G, Blanco E, Calvete C, Cavadini P, Cooke B, Dalton K, Delibes Mateos M, Deptula W, Eden JS, Wang F, Ferreira CC, Ferreira P, Foronda P, Goncalves D, Gavier-Widen D, Hall R, Hukowska-Szematowicz B, Kerr P, Kovaliski J, Lavazza A, Mahar J, Malogolovkin A, Marques RM, Marques S, Martin-Alonso A, Monterroso P, Moreno S, Mutze G, Neimanis A, Niedzwiedzka-Rystwej P, Peacock D, Parra F, Rocchi M, Rouco C, Ruvoen-Clouet N, Silva E, Silverio D, Strive T, Thompson G, Tokarz-Deptula B, Esteves P. 2017. Proposal for a unified classification system and nomenclature of lagoviruses. J Gen Virol 98:1658–1666. doi: 10.1099/jgv.0.000840. - DOI - PubMed
-
- Duarte M, Carvalho C, Bernardo S, Barros SV, Benevides S, Flor L, Monteiro M, Marques I, Henriques M, Barros SC, Fagulha T, Ramos F, Luis T, Fevereiro M. 2015. Rabbit haemorrhagic disease virus 2 (RHDV2) outbreak in Azores: disclosure of common genetic markers and phylogenetic segregation within the European strains. Infect Genet Evol 35:163–171. doi: 10.1016/j.meegid.2015.08.005. - DOI - PubMed
-
- Le Gall-Recule G, Lavazza A, Marchandeau S, Bertagnoli S, Zwingelstein F, Cavadini P, Martinelli N, Lombardi G, Guerin JL, Lemaitre E, Decors A, Boucher S, Le Normand B, Capucci L. 2013. Emergence of a new lagovirus related to rabbit haemorrhagic disease virus. Vet Res 44:81. doi: 10.1186/1297-9716-44-81. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

