Inducible MicroRNA-3570 Feedback Inhibits the RIG-I-Dependent Innate Immune Response to Rhabdovirus in Teleost Fish by Targeting MAVS/IPS-1
- PMID: 29093090
- PMCID: PMC5752954
- DOI: 10.1128/JVI.01594-17
Inducible MicroRNA-3570 Feedback Inhibits the RIG-I-Dependent Innate Immune Response to Rhabdovirus in Teleost Fish by Targeting MAVS/IPS-1
Abstract
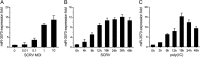
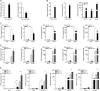
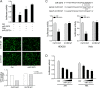
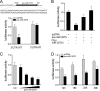
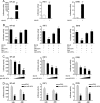
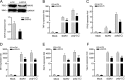
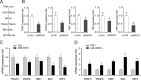
Effectively recognizing invading viruses and subsequently inducing innate antiviral immunity are essential for host antiviral defense. Although these processes are closely regulated by the host to maintain immune balance, viruses have evolved the ability to downregulate or upregulate these processes for their survival. MicroRNAs (miRNAs) are a family of small noncoding RNAs that play vital roles in modulating host immune response. Accumulating evidence demonstrates that host miRNAs as mediators are involved in regulating viral replication and host antiviral immunity in mammals. However, the underlying regulatory mechanisms in fish species are still poorly understood. Here, we found that rhabdovirus infection significantly upregulated host miR-3570 expression in miiuy croaker macrophages. Induced miR-3570 negatively modulated RNA virus-triggered type I interferon (IFN) and antiviral gene production, thus facilitating viral replication. Furthermore, miR-3570 was found to target and posttranscriptionally downregulate mitochondrial antiviral signaling protein (MAVS), which functions as a platform for innate antiviral signal transduction. Moreover, we demonstrated that miR-3570 suppressed the expression of MAVS, thereby inhibiting MAVS-mediated NF-κB and IRF3 signaling. The collective results demonstrated a novel regulation mechanism of MAVS-mediated immunity during RNA viral infection by miRNA.IMPORTANCE RNA viral infection could upregulate host miR-3570 expression in miiuy croaker macrophages. Induced miR-3570 negatively modulates RNA virus-triggered type I IFN and antiviral gene production, thus facilitating viral replication. Remarkably, miR-3570 could target and inhibit MAVS expression, which thus modulates MAVS-mediated NF-κB and IRF3 signaling. The collective results of this study suggest a novel regulation mechanism of MAVS-mediated immunity during RNA viral infection by miR-3570. Thus, a novel mechanism for virus evasion in fish is proposed.
Keywords: MAVS; miR-3570; microRNA; teleost fish; virus evasion.
Copyright © 2018 American Society for Microbiology.
Figures

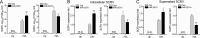

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

