Canalization of Tomato Fruit Metabolism
- PMID: 29093214
- PMCID: PMC5728129
- DOI: 10.1105/tpc.17.00367
Canalization of Tomato Fruit Metabolism
Abstract
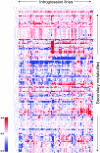
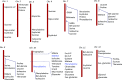
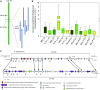
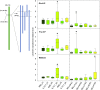
To explore the genetic robustness (canalization) of metabolism, we examined the levels of fruit metabolites in multiple harvests of a tomato introgression line (IL) population. The IL partitions the whole genome of the wild species Solanum pennellii in the background of the cultivated tomato (Solanum lycopersicum). We identified several metabolite quantitative trait loci that reduce variability for both primary and secondary metabolites, which we named canalization metabolite quantitative trait loci (cmQTL). We validated nine cmQTL using an independent population of backcross inbred lines, derived from the same parents, which allows increased resolution in mapping the QTL previously identified in the ILs. These cmQTL showed little overlap with QTL for the metabolite levels themselves. Moreover, the intervals they mapped to harbored few metabolism-associated genes, suggesting that the canalization of metabolism is largely controlled by regulatory genes.
© 2017 American Society of Plant Biologists. All rights reserved.
Figures


References
-
- Alon U., Surette M.G., Barkai N., Leibler S. (1999). Robustness in bacterial chemotaxis. Nature 397: 168–171. - PubMed
-
- Becker H.C., Leon J. (1988). Stability analysis in plant-breeding. Plant Breed. 101: 1–23.
-
- Capel C., Yuste-Lisbona F.J., López-Casado G., Angosto T., Cuartero J., Lozano R., Capel J. (2017a). Multi-environment QTL mapping reveals genetic architecture of fruit cracking in a tomato RIL Solanum lycopersicum × S. pimpinellifolium population. Theor. Appl. Genet. 130: 213–222. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

