HtrA1 activation is driven by an allosteric mechanism of inter-monomer communication
- PMID: 29093542
- PMCID: PMC5666011
- DOI: 10.1038/s41598-017-14208-z
HtrA1 activation is driven by an allosteric mechanism of inter-monomer communication
Abstract
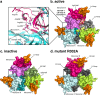
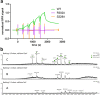
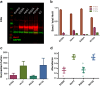
The human protease family HtrA is responsible for preventing protein misfolding and mislocalization, and a key player in several cellular processes. Among these, HtrA1 is implicated in several cancers, cerebrovascular disease and age-related macular degeneration. Currently, HtrA1 activation is not fully characterized and relevant for drug-targeting this protease. Our work provides a mechanistic step-by-step description of HtrA1 activation and regulation. We report that the HtrA1 trimer is regulated by an allosteric mechanism by which monomers relay the activation signal to each other, in a PDZ-domain independent fashion. Notably, we show that inhibitor binding is precluded if HtrA1 monomers cannot communicate with each other. Our study establishes how HtrA1 trimerization plays a fundamental role in proteolytic activity. Moreover, it offers a structural explanation for HtrA1-defective pathologies as well as mechanistic insights into the degradation of complex extracellular fibrils such as tubulin, amyloid beta and tau that belong to the repertoire of HtrA1.
Conflict of interest statement
All authors are F. Hoffman-La Roche employees or collaborators.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

