Epstein-Barr virus-associated primary nodal T/NK-cell lymphoma shows a distinct molecular signature and copy number changes
- PMID: 29097495
- PMCID: PMC5792272
- DOI: 10.3324/haematol.2017.180430
Epstein-Barr virus-associated primary nodal T/NK-cell lymphoma shows a distinct molecular signature and copy number changes
Abstract
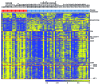
The molecular biology of primary nodal T- and NK-cell lymphoma and its relationship with extranodal NK/T-cell lymphoma, nasal type is poorly understood. In this study, we assessed the relationship between nodal and extranodal Epstein-Barr virus-positive T/NK-cell lymphomas using gene expression profiling and copy number aberration analyses. We performed gene expression profiling and copy number aberration analysis on 66 cases of Epstein-Barr virus-associated T/NK-cell lymphoma from nodal and extranodal sites, and correlated the molecular signatures with clinicopathological features. Three distinct molecular clusters were identified with one enriched for nodal presentation and loss of 14q11.2 (TCRA loci). T/NK-cell lymphomas with a nodal presentation (nodal-group) were significantly associated with older age, lack of nasal involvement, and T-cell lineage compared to those with an extranodal presentation (extranodal-group). On multivariate analysis, nodal presentation was an independent factor associated with short survival. Comparing the molecular signatures of the nodal and extranodal groups it was seen that the former was characterized by upregulation of PD-L1 and T-cell-related genes, including CD2 and CD8, and downregulation of CD56, consistent with the CD8+/CD56-immunophenotype. PD-L1 and CD2 protein expression levels were validated using multiplexed immunofluorescence. Interestingly, nodal group lymphomas were associated with 14q11.2 loss which correlated with loss of TCR loci and T-cell origin. Overall, our results suggest that T/NK-cell lymphoma with nodal presentation is distinct and deserves to be classified separately from T/NK-cell lymphoma with extranodal presentation. Upregulation of PD-L1 indicates that it may be possible to use anti-PD1 immunotherapy in this distinctive entity. In addition, loss of 14q11.2 may be a potentially useful diagnostic marker of T-cell lineage.
Copyright© 2018 Ferrata Storti Foundation.
Figures




References
-
- Attygalle AD, Cabecadas J, Gaulard P, et al. Peripheral T-cell and NK-cell lymphomas and their mimics; taking a step forward -report on the lymphoma workshop of the XVIth meeting of the European Association for Haematopathology and the Society for Hematopathology. Histopathology. 2014;64(2):171–199. - PMC - PubMed
-
- Chan JKC, Quintanilla-Martinez L, Ferry JA, Peh SC. Extranodal NK/T-cell lymphoma, nasal type. In: Swerdlow SH, Campo E, Harris NL, et al., eds. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: IRAC Press; 2008:285–288.
-
- Ko YH, Chan JKC, Quintanilla-Martinez L. Virally associated T-cell and NK-cell Neoplasms. In: Jaffe ES, Arber DA, Campo E, Harris NL, Quintanilla-Martinez L, eds. Hematopathology. Philadelphia: Elsevier; 2017:565–598.
-
- Kato S, Asano N, Miyata-Takata T, et al. T-cell receptor (TCR) phenotype of nodal Epstein-Barr virus (EBV)-positive cytotoxic T-cell lymphoma (CTL): a clinicopathologic study of 39 cases. Am J Surg Pathol. 2015;39(4):462–471. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

