The genetic variation in the R1a clade among the Ashkenazi Levites' Y chromosome
- PMID: 29097670
- PMCID: PMC5668307
- DOI: 10.1038/s41598-017-14761-7
The genetic variation in the R1a clade among the Ashkenazi Levites' Y chromosome
Abstract
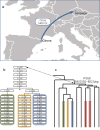
Approximately 300,000 men around the globe self-identify as Ashkenazi Levites, of whom two thirds were previously shown to descend from a single male. The paucity of whole Y-chromosome sequences precluded conclusive identification of this ancestor's age, geographic origin and migration patterns. Here, we report the variation of 486 Y-chromosomes within the Ashkenazi and non-Ashkenazi Levite R1a clade, other Ashkenazi Jewish paternal lineages, as well as non-Levite Jewish and non-Jewish R1a samples. Cumulatively, the emerging profile is of a Middle Eastern ancestor, self-affiliating as Levite, and carrying the highly resolved R1a-Y2619 lineage, which was likely a minor haplogroup among the Hebrews. A star-like phylogeny, coalescing similarly to other Ashkenazi paternal lineages, ~1,743 ybp, suggests it to be one of the Ashkenazi paternal founders; to have expanded as part of the overall Ashkenazi demographic expansion, without special relation to the Levite affiliation; and to have subsequently spread to non-Ashkenazi Levites.
Conflict of interest statement
Part of the lab work presented in this study was conducted in Gene by Gene (Family Tree DNA) in which Doron M. Behar, Elliott Greenspan and Concetta Bormans declare stock ownership and Luisa Fernanda Sanchez is an employee. The other authors claim no competing financial interests associated with this paper.
Figures


References
-
- Hey, D. family Names and Family History. (Hambledon & London, 2006).
-
- Jobling MA, Tyler-Smith C. Human Y-chromosome variation in the genome-sequencing era. Nature reviews. Genetics. 2017 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

