Identification of miRNA and genes involving in osteosarcoma by comprehensive analysis of microRNA and copy number variation data
- PMID: 29098032
- PMCID: PMC5652194
- DOI: 10.3892/ol.2017.6845
Identification of miRNA and genes involving in osteosarcoma by comprehensive analysis of microRNA and copy number variation data
Abstract
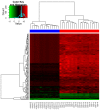
The aim of the present study was to understand the molecular mechanisms of osteosarcoma by comprehensive analysis of microRNA (miRNA/miR) and copy number variation (CNV) microarray data. Microarray data (GSE65071 and GSE33153) were downloaded from the Gene Expression Omnibus. In GSE65071, differentially expressed miRNAs between the osteosarcoma and control groups were calculated by the Limma package. Target genes of differentially expressed miRNAs were identified by the starBase database. For GSE33153, PennCNV software was used to perform the copy number variation (CNV) analysis. Overlapping of the genes in CNV regions and the target genes of differentially expressed miRNAs were used to construct miRNA-gene regulatory network using the starBase database. A total of 149 differentially expressed miRNAs, including 13 downregulated and 136 upregulated, were identified. In the GSE33153 dataset, 987 CNV regions involving in 3,635 genes were identified. In total, 761 overlapping genes in 987 CNV regions and in the genes in 7,313 miRNA-gene pairs were obtained. miRNAs (hsa-miR-27a-3p, hsa-miR-124-3p, hsa-miR-9-5p, hsa-miR-182-5p, hsa-miR-26a-5p) and the genes [Fibroblast growth factor receptor substrate 2 (FRS2), coronin 1C (CORO1C), forkhead box P1 (FOXP1), cytoplasmic polyadenylation element binding protein 4 (CPEB4) and glucocorticoid induced 1 (GLCCI1)] with the highest degrees of association with osteosarcoma development were identified. Hsa-miR-27a-3p, hsa-miR-9-5p, hsa-miR-182-5p, FRS2, CORO1C, FOXP1 and CPEB4 may be involved in osteosarcoma pathogenesis, and development.
Keywords: copy number variation; microRNA; microRNA-gene regulatory network; osteosarcoma.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
