Atypical fibroxanthoma and pleomorphic dermal sarcoma harbor frequent NOTCH1/2 and FAT1 mutations and similar DNA copy number alteration profiles
- PMID: 29099504
- PMCID: PMC7463132
- DOI: 10.1038/modpathol.2017.146
Atypical fibroxanthoma and pleomorphic dermal sarcoma harbor frequent NOTCH1/2 and FAT1 mutations and similar DNA copy number alteration profiles
Abstract
Atypical fibroxanthomas and pleomorphic dermal sarcomas are tumors arising in sun-damaged skin of elderly patients. They have differing prognoses and are currently distinguished using histological criteria, such as invasion of deeper tissue structures, necrosis and lymphovascular or perineural invasion. To investigate the as-yet poorly understood genetics of these tumors, 41 atypical fibroxanthomas and 40 pleomorphic dermal sarcomas were subjected to targeted next-generation sequencing approaches as well as DNA copy number analysis by comparative genomic hybridization. In an analysis of the entire coding region of 341 oncogenes and tumor suppressor genes in 13 atypical fibroxanthomas using an established hybridization-based next-generation sequencing approach, we found that these tumors harbor a large number of mutations. Gene alterations were identified in more than half of the analyzed samples in FAT1, NOTCH1/2, CDKN2A, TP53, and the TERT promoter. The presence of these alterations was verified in 26 atypical fibroxanthoma and 35 pleomorphic dermal sarcoma samples by targeted amplicon-based next-generation sequencing. Similar mutation profiles in FAT1, NOTCH1/2, CDKN2A, TP53, and the TERT promoter were identified in both atypical fibroxanthoma and pleomorphic dermal sarcoma. Activating RAS mutations (G12 and G13) identified in 3 pleomorphic dermal sarcoma were not found in atypical fibroxanthoma. Comprehensive DNA copy number analysis demonstrated a wide array of different copy number gains and losses, with similar profiles in atypical fibroxanthoma and pleomorphic dermal sarcoma. In summary, atypical fibroxanthoma and pleomorphic dermal sarcoma are highly mutated tumors with recurrent mutations in FAT1, NOTCH1/2, CDKN2A, TP53, and the TERT promoter, and a range of DNA copy number alterations. These findings suggest that atypical fibroxanthomas and pleomorphic dermal sarcomas are genetically related, potentially representing two ends of a common tumor spectrum and distinguishing these entities is at present still best performed using histological criteria.
Conflict of interest statement
Disclosure/conflict of interest
The authors declare no conflict of interest.
Figures
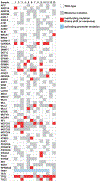


References
-
- Iorizzo LJ 3rd, Brown MD. Atypical fibroxanthoma: a review of the literature. Dermatol Surg 2011;37: 146–157. - PubMed
-
- Gru AA, Santa Cruz DJ. Atypical fibroxanthoma: a selective review. Semin Diagn Pathol 2013;30:4–12. - PubMed
-
- McCalmont TH. AFX: what we now know. J Cutan Pathol 2011;38:853–856. - PubMed
-
- Mentzel T, Requena L, Brenn T. Atypical fibroxanthoma revisited. Surg Pathol Clin 2017;10:319–335. - PubMed
-
- McCoppin HH, Christiansen D, Stasko T, et al. Clinical spectrum of atypical fibroxanthoma and undifferentiated pleomorphic sarcoma in solid organ transplant recipients: a collective experience. Dermatol Surg 2012;38:230–239. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

