Genome-wide analysis of MATE transporters and molecular characterization of aluminum resistance in Populus
- PMID: 29099944
- PMCID: PMC5853298
- DOI: 10.1093/jxb/erx370
Genome-wide analysis of MATE transporters and molecular characterization of aluminum resistance in Populus
Abstract
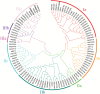
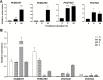
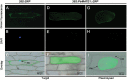
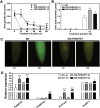
Ionic aluminum (Al) in acidic soils, comprising approximately 50% of arable land globally, is highly toxic to most plant species. Populus grow naturally in acidic soils and tolerate high concentrations of Al. Multidrug and toxic compound extrusion (MATE) family genes in plants are involved in responses to Al tolerance. To date, however, the functional roles of the MATE genes in Populus remain unclear. In the present study, 71 putative MATE transporters were predicted in the genome of Populus trichocarpa. The chromosome distribution, phylogenetic relationships, and expression level analysis revealed that four candidate MATE genes belonging to subgroup IIIc might contribute to high Al tolerance in poplar. Further, the expression levels of two subgroup IIIc members, PtrMATE1 and PtrMATE2, were induced by Al stress. Transient expression in onion epidermal cells showed that PtrMATE1 was localized to the plasma membrane. Overexpression of PtrMATE1 increased Al-induced secretion of citrate from the root apex of transgenic plants. Al-induced inhibition of root growths were alleviated in both PtrMATE1 overexpression lines in Populus and in Arabidopsis compared with wild-type plants. In addition, PtrMATE1 expression was induced at 12 h after exposure to Al stress whereas PtrMATE2 expression was induced at 24 h, indicating that these proteins coordinately function in response to Al stress in poplar. Taken together, these results provide important insights into the molecular mechanisms involved in Al tolerance in poplar.
Keywords: Aluminum stress; MATE transporter; Populus; citrate exudation; root apex.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures




References
-
- Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings. International Conference on Intelligent Systems for Molecular Biology 2, 28–36. - PubMed
-
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

