De Novo Mutations in Protein Kinase Genes CAMK2A and CAMK2B Cause Intellectual Disability
- PMID: 29100089
- PMCID: PMC5673671
- DOI: 10.1016/j.ajhg.2017.10.003
De Novo Mutations in Protein Kinase Genes CAMK2A and CAMK2B Cause Intellectual Disability
Abstract
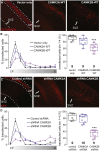
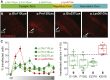
Calcium/calmodulin-dependent protein kinase II (CAMK2) is one of the first proteins shown to be essential for normal learning and synaptic plasticity in mice, but its requirement for human brain development has not yet been established. Through a multi-center collaborative study based on a whole-exome sequencing approach, we identified 19 exceedingly rare de novo CAMK2A or CAMK2B variants in 24 unrelated individuals with intellectual disability. Variants were assessed for their effect on CAMK2 function and on neuronal migration. For both CAMK2A and CAMK2B, we identified mutations that decreased or increased CAMK2 auto-phosphorylation at Thr286/Thr287. We further found that all mutations affecting auto-phosphorylation also affected neuronal migration, highlighting the importance of tightly regulated CAMK2 auto-phosphorylation in neuronal function and neurodevelopment. Our data establish the importance of CAMK2A and CAMK2B and their auto-phosphorylation in human brain function and expand the phenotypic spectrum of the disorders caused by variants in key players of the glutamatergic signaling pathway.
Keywords: AMPAR; CAMK2; CAMK2A; CAMK2B; NMDAR; de novo mutations; intellectual disability; synaptic plasticity.
Copyright © 2017 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

