Plasma exosome miR-196a and miR-1246 are potential indicators of localized pancreatic cancer
- PMID: 29100367
- PMCID: PMC5652761
- DOI: 10.18632/oncotarget.20332
Plasma exosome miR-196a and miR-1246 are potential indicators of localized pancreatic cancer
Abstract
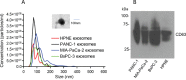
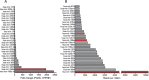
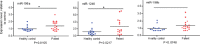
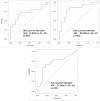
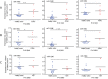
Patients with localized pancreatic cancer (stage I and stage IIA) have a much higher survival rate than those presenting at later stages, yet early detection remains a challenge to this malignancy. The aim of this study was to evaluate whether exosome miRNA signatures are indicative of localized pancreatic cancer. Exosomes were collected from the conditioned media of pancreatic cancer cell lines and plasma samples of localized pancreatic cancer patients (Stage I-IIA, n=15), and healthy subjects (n=15). Cellular and exosome miRNAs from pancreatic cancer cell lines were profiled by next-generation small RNA sequencing. Plasma exosome miRNA expression was analyzed by qRT-PCR. We found that certain miRNAs, such as miR-196a and miR-1246, are highly enriched in pancreatic cancer exosomes. Consistently, plasma exosome miR-196a and miR-1246 levels were significantly elevated in pancreatic cancer patients as compared to healthy subjects. An analysis of the cancer subtypes indicated that plasma exosome miR-196a is a better indicator of pancreatic ductal adenocarcinoma (PDAC), whereas plasma exosome miR-1246 is significantly elevated in patients with intraductal papillary mucinous neoplasms (IPMN). In contrast, there were no differences in the plasma exosome miR-196a and miR-1246 levels between patients with pancreatic neuroendocrine tumors (NET) and healthy subjects. In conclusion, we demonstrate that certain miRNA species, such as miR-196a and miR-1246, are highly enriched in pancreatic cancer exosomes and elevated in plasma exosomes of patients with localized pancreatic cancer.
Keywords: early detection; exosome; microRNAs; pancreatic cancer; plasma biomarkers.
Conflict of interest statement
CONFLICTS OF INTEREST There are no conflicts to disclose.
Figures
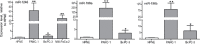

References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. - PubMed
-
- NATIONAL CANCER INSTITUTE-SEER. Stat Fact Sheets: Pancreas Cancer. http://seer.cancer.gov/statfacts/html/pancreas.html Accessed on 8 March, 2017.
-
- NATIONAL CANCER. INSTITUTE--SEER Stat Fact Sheets: All Cancer Sites. http://seer.cancer.gov/statfacts/html/all.html Accessed on 8 March, 2017.
-
- Hanada K, Okazaki A, Hirano N, Izumi Y, Minami T, Ikemoto J, Kanemitsu K, Hino F. Effective screening for early diagnosis of pancreatic cancer. Best practice & research Clinical gastroenterology. 2015;29:929–939. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

