Targeting Hsp27/eIF4E interaction with phenazine compound: a promising alternative for castration-resistant prostate cancer treatment
- PMID: 29100389
- PMCID: PMC5652782
- DOI: 10.18632/oncotarget.20469
Targeting Hsp27/eIF4E interaction with phenazine compound: a promising alternative for castration-resistant prostate cancer treatment
Erratum in
-
Correction: Targeting Hsp27/eIF4E interaction with phenazine compound: a promising alternative for castration-resistant prostate cancer treatment.Oncotarget. 2018 Jun 19;9(47):28797. doi: 10.18632/oncotarget.25683. eCollection 2018 Jun 19. Oncotarget. 2018. PMID: 29983898 Free PMC article.
Abstract
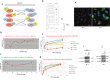
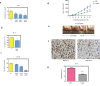
The actual strategy to improve current therapies in advanced prostate cancer involves targeting genes activated by androgen withdrawal, either to delay or prevent the emergence of the castration-refractory phenotype. However, these genes are often implicated in several physiological processes, and long-term inhibition of survival proteins might be accompanied with cytotoxic effects. To avoid this problem, an alternative therapeutic strategy relies on the identification and use of compounds that disrupt specific protein-protein interactions involved in androgen withdrawal. Specifically, the interaction of the chaperone protein Hsp27 with the initiation factor eIF4E leads to the protection of protein synthesis initiation process and enhances cell survival during cell stress induced by castration or chemotherapy. Thus, in this work we aimed at i) identifying the interaction site of the Hsp27/eIF4E complex and ii) interfere with the relevant protein/protein association mechanism involved in castration-resistant progression of prostate cancer. By a combination of experimental and modeling techniques, we proved that eIF4E interacts with the C-terminal part of Hsp27, preferentially when Hsp27 is phosphorylated. We also observed that the loss of this interaction increased cell chemo-and hormone-sensitivity. In order to find a potential inhibitor of Hsp27/eIF4E interaction, BRET assays in combination with molecular simulations identified the phenazine derivative 14 as the compound able to efficiently interfere with this protein/protein interaction, thereby inhibiting cell viability and increasing cell death in chemo- and castration-resistant prostate cancer models in vitro and in vivo.
Keywords: Hsp27/eIF4E interaction; prostate cancer; protein-protein interaction inhibition.
Conflict of interest statement
CONFLICTS OF INTEREST The authors declare no conflicts of interest.
Figures


References
-
- Resnick MJ, Lacchetti C, Penson DF. Prostate cancer survivorship care guidelines: American Society of Clinical Oncology practice guideline endorsement. J Oncol Pract. 2015;11:e445–449. - PubMed
-
- Badrising S, van der Noort V, van Oort IM, van den Berg HP, Los M, Hamberg P, Coenen JL, van den Eertwegh AJ, de Jong IJ, Kerver ED, van Tinteren H, Bergman AM. Clinical activity and tolerability of enzalutamide (MDV3100) in patients with metastatic, castration-resistant prostate cancer who progress after docetaxel and abiraterone treatment. Cancer. 2014;120:968–975. - PubMed
-
- Katsogiannou M, Ziouziou H, Karaki S, Andrieu C, Henry de Villeneuve M, Rocchi P. The hallmarks of castration-resistant prostate cancers. Cancer Treat Rev. 2015;41:588–597. - PubMed
-
- Mymrikov EV, Seit-Nebi AS, Gusev NB. Large potentials of small heat shock proteins. Physiol Rev. 2011;91:1123–1159. - PubMed
-
- Acunzo J, Andrieu C, Baylot V, So A, Rocchi P. Hsp27 as a therapeutic target in cancers. Curr Drug Targets. 2014;15:423–431. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

