Gene-body hypermethylation controlled cryptic promoter and miR26A1-dependent EZH2 regulation of TET1 gene activity in chronic lymphocytic leukemia
- PMID: 29100411
- PMCID: PMC5652802
- DOI: 10.18632/oncotarget.20668
Gene-body hypermethylation controlled cryptic promoter and miR26A1-dependent EZH2 regulation of TET1 gene activity in chronic lymphocytic leukemia
Abstract
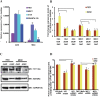
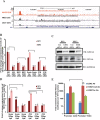
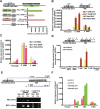
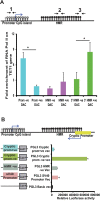
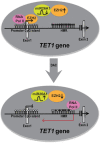
The Ten-eleven-translocation 1 (TET1) protein is a member of dioxygenase protein family that catalyzes the oxidation of 5-methylcytosine to 5-hydroxymethylcytosine. TET1 is differentially expressed in many cancers, including leukemia. However, very little is known about mechanism behind TET1 deregulation. Previously, by characterizing global methylation patterns in CLL patients using MBD-seq, we found TET1 as one of the differentially methylated regions with gene-body hypermethylation. Herein, we characterize mechanisms that control TET1 gene activity at the transcriptional level. We show that treatment of CLL cell lines with 5-aza 2´-deoxycytidine (DAC) results in the activation of miR26A1, which causes decrease in both mRNA and protein levels of EZH2, which in turn results in the decreased occupancy of EZH2 over the TET1 promoter and consequently the loss of TET1 expression. In addition, DAC treatment also leads to the activation of antisense transcription overlapping the TET1 gene from a cryptic promoter, located in the hypermethylated intronic region. Increased expression of intronic transcripts correlates with decreased TET1 promoter activity through the loss of RNA Pol II occupancy. Thus, our data demonstrate that TET1 gene activation in CLL depends on miR26A1 regulated EZH2 binding at the TET1 promoter and silencing of novel cryptic promoter by gene-body hypermethylation.
Keywords: EZH2 and TET1 gene; chronic lymphocytic leukemia; cryptic promoters; gene-body hypermethylation.
Conflict of interest statement
CONFLICTS OF INTEREST The authors have no conflicts of interest.
Figures

Similar articles
-
Epigenetic silencing of miR-26A1 in chronic lymphocytic leukemia and mantle cell lymphoma: Impact on EZH2 expression.Epigenetics. 2016 May 3;11(5):335-43. doi: 10.1080/15592294.2016.1164375. Epub 2016 Apr 6. Epigenetics. 2016. PMID: 27052808 Free PMC article.
-
Loss of TET1 facilitates DLD1 colon cancer cell migration via H3K27me3-mediated down-regulation of E-cadherin.J Cell Physiol. 2018 Feb;233(2):1359-1369. doi: 10.1002/jcp.26012. Epub 2017 Jun 12. J Cell Physiol. 2018. PMID: 28513825
-
Aberrant methylation and silencing of ARHI, an imprinted tumor suppressor gene in which the function is lost in breast cancers.Cancer Res. 2003 Jul 15;63(14):4174-80. Cancer Res. 2003. PMID: 12874023
-
Decrease of 5hmC in gastric cancers is associated with TET1 silencing due to with DNA methylation and bivalent histone marks at TET1 CpG island 3'-shore.Oncotarget. 2015 Nov 10;6(35):37647-62. doi: 10.18632/oncotarget.6069. Oncotarget. 2015. PMID: 26462176 Free PMC article.
-
Multiple Functions of Ten-eleven Translocation 1 during Tumorigenesis.Chin Med J (Engl). 2016 Jul 20;129(14):1744-51. doi: 10.4103/0366-6999.185873. Chin Med J (Engl). 2016. PMID: 27411465 Free PMC article. Review.
Cited by
-
DNA hypermethylation preceded by H3K27 trimethylation is linked to downregulation of gene expression in disuse muscle atrophy in male mice.Physiol Rep. 2025 Apr;13(7):e70317. doi: 10.14814/phy2.70317. Physiol Rep. 2025. PMID: 40223401 Free PMC article.
-
Decitabine-induced DNA methylation-mediated transcriptomic reprogramming in human breast cancer cell lines; the impact of DCK overexpression.Front Pharmacol. 2022 Oct 5;13:991751. doi: 10.3389/fphar.2022.991751. eCollection 2022. Front Pharmacol. 2022. PMID: 36278182 Free PMC article.
-
The Role of Methylation in Chronic Lymphocytic Leukemia and Its Prognostic and Therapeutic Impacts in the Disease: A Systematic Review.Adv Hematol. 2024 Feb 23;2024:1370364. doi: 10.1155/2024/1370364. eCollection 2024. Adv Hematol. 2024. PMID: 38435839 Free PMC article. Review.
-
Global distribution of DNA hydroxymethylation and DNA methylation in chronic lymphocytic leukemia.Epigenetics Chromatin. 2019 Jan 7;12(1):4. doi: 10.1186/s13072-018-0252-7. Epigenetics Chromatin. 2019. PMID: 30616658 Free PMC article.
References
-
- Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RA, Kamping EJ, Verhoef GE, Verburgh E, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41:838–842. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

