Kinetic modeling of Shewanella baltica KB30 growth on different substrates through respirometry
- PMID: 29100519
- PMCID: PMC5670636
- DOI: 10.1186/s12934-017-0805-7
Kinetic modeling of Shewanella baltica KB30 growth on different substrates through respirometry
Abstract
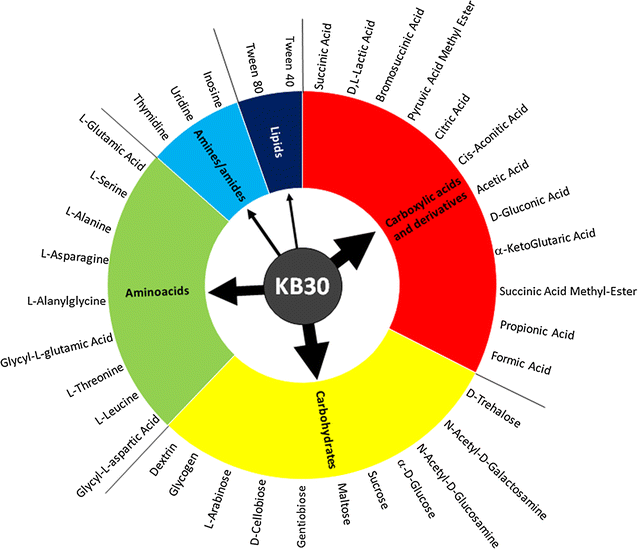
Background: Shewanella baltica KB30 was isolated from seawater collected in Kandalaksha Bay, White Sea (Russia). This strain is known for its ability to grow on a pool of different substrates, including carbohydrates, carboxylic and amino acids, and lipids. However, no data are available on its metabolic efficiency in relation to the use of different carbon sources typologies. This work represents the first attempt to characterize S. baltica by its heterotrophic kinetic performance.
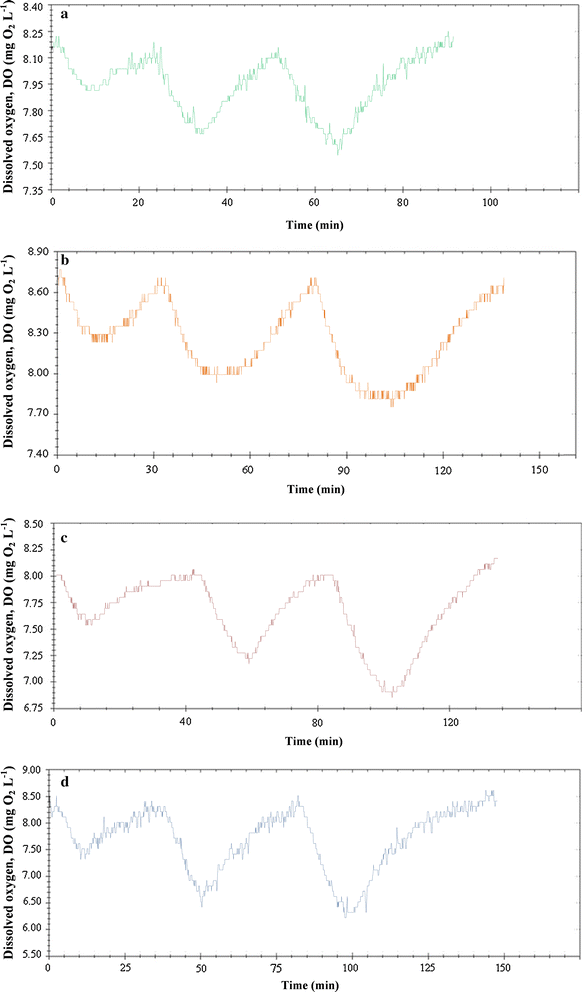
Results: Growth and substrate consumption, during the biodegradation of sodium acetate, glucose, tween 80 and peptone, were analyzed through a respirometric method. To find the model best fitting the experimental data and to obtain the kinetic parameters, the equations of Monod, Moser, Contois and Tessier were applied. The kinetic behavior of S. baltica was fitted to Monod model for sodium acetate and tween 80, while it was adjusted to Contois model for glucose and peptone. In this regard, peptone was consumed faster than the other substrates, as indicated by the highest values of substrate degradation rate, which exceeded 60 mg O2 L-1 h-1.
Conclusions: Proteolytic metabolism was favored than lipidic and glucidic metabolism, which could contribute much more to mineralization and recycling of proteins than lipids and carbohydrates.
Keywords: Carbon utilization; Kinetics; Modeling; Shewanella baltica.
Figures



Similar articles
-
Influence of acetate and CO2 on the TMAO-reduction reaction by Shewanella baltica.Int J Food Microbiol. 2001 Aug 15;68(1-2):115-23. doi: 10.1016/s0168-1605(01)00484-6. Int J Food Microbiol. 2001. PMID: 11545211
-
Identification of Shewanella baltica as the most important H2S-producing species during iced storage of Danish marine fish.Appl Environ Microbiol. 2005 Nov;71(11):6689-97. doi: 10.1128/AEM.71.11.6689-6697.2005. Appl Environ Microbiol. 2005. PMID: 16269698 Free PMC article.
-
Tellurite biotransformation and detoxification by Shewanella baltica with simultaneous synthesis of tellurium nanorods exhibiting photo-catalytic and anti-biofilm activity.Ecotoxicol Environ Saf. 2018 Dec 15;165:516-526. doi: 10.1016/j.ecoenv.2018.08.111. Epub 2018 Sep 14. Ecotoxicol Environ Saf. 2018. PMID: 30223164
-
Shewanella hafniensis sp. nov. and Shewanella morhuae sp. nov., isolated from marine fish of the Baltic Sea.Int J Syst Evol Microbiol. 2006 Jan;56(Pt 1):243-9. doi: 10.1099/ijs.0.63931-0. Int J Syst Evol Microbiol. 2006. PMID: 16403893
-
Modeling aerobic carbon source degradation processes using titrimetric data and combined respirometric-titrimetric data: structural and practical identifiability.Biotechnol Bioeng. 2002 Sep 30;79(7):754-67. doi: 10.1002/bit.10337. Biotechnol Bioeng. 2002. PMID: 12209798
Cited by
-
Extended antibiotic treatment in salmon farms select multiresistant gut bacteria with a high prevalence of antibiotic resistance genes.PLoS One. 2018 Sep 11;13(9):e0203641. doi: 10.1371/journal.pone.0203641. eCollection 2018. PLoS One. 2018. PMID: 30204782 Free PMC article.
-
Impact of Fe(III) (Oxyhydr)oxides Mineralogy on Iron Solubilization and Associated Microbial Communities.Front Microbiol. 2020 Nov 20;11:571244. doi: 10.3389/fmicb.2020.571244. eCollection 2020. Front Microbiol. 2020. PMID: 33329429 Free PMC article.
References
-
- Shewan JM, Hobbs G, Hodgkiss W. A determinative scheme for the identification of certain genera of Gram-negative bacteria with special reference to Pseudomonodaceae. J Appl Bacteriol. 1960;23:379–390. doi: 10.1111/j.1365-2672.1960.tb00211.x. - DOI
-
- Macdonell MT, Colwell RR. Phylogeny of the Vibrionaceae and recommendation for two new genera Listonella and Shewanella. Syst Appl Microbiol. 1985;6:171–182. doi: 10.1016/S0723-2020(85)80051-5. - DOI
-
- Ivanova EP, Flavier S, Christen R. Phylogenetic relationships among marine Alteromonas-like proteobacteria: emended description of the family Alteromonadaceae and proposal of Pseudoalteromonadaceae fam. nov., Colwelliaceae fam. nov., Shewanellaceae fam. nov., Moritellaceae fam. nov., Ferrimonadaceae fam. nov., Idiomarinaceae fam. nov. and Psychromonadaceae fam. nov. Int J Syst Evol Microbiol. 2004;54(5):1773–1788. doi: 10.1099/ijs.0.02997-0. - DOI - PubMed
-
- Bowman JP, McCammon SA, Nichols DS, Skerratt JH, Rea SM, Nichols PD, McMeekin TA. Shewanella gelidimarina sp. nov. and Shewanella frigidimarina sp. nov., novel Antarctic species with the ability to produce eicosapentaenoic acid (20:5w3) and grow anaerobically by dissimilatory Fe(III) reduction. Int J Syst Bacteriol. 1997;47:1040–1047. doi: 10.1099/00207713-47-4-1040. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

