Construction and characterization of an improved DNA-launched infectious clone of duck hepatitis a virus type 1
- PMID: 29100535
- PMCID: PMC5670519
- DOI: 10.1186/s12985-017-0883-5
Construction and characterization of an improved DNA-launched infectious clone of duck hepatitis a virus type 1
Abstract
Background: DNA-launched infectious system is a useful tool with high rescue efficiency that allows the introduction of mutations in specific positions to investigate the function of an individual viral element. Rescued virus particles could be harvested by directly transfecting the DNA-launched recombinant plasmid to the host cells, which will reduce labor and experimental cost by skipping the in vitro transcription assay.
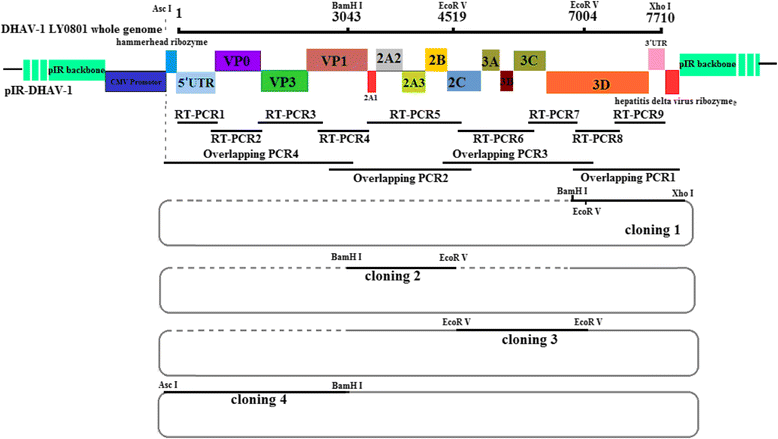
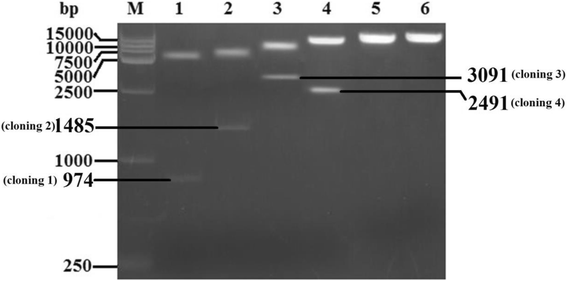
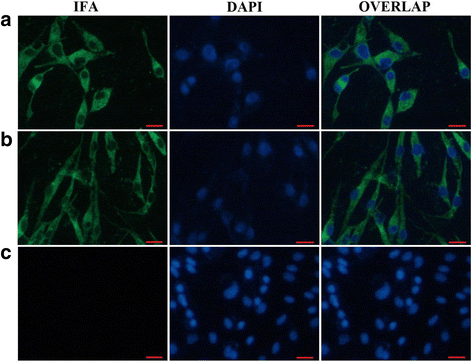
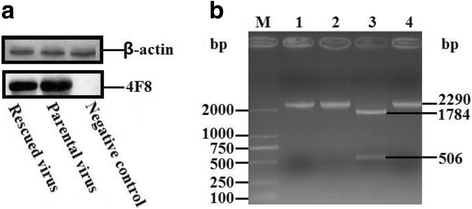
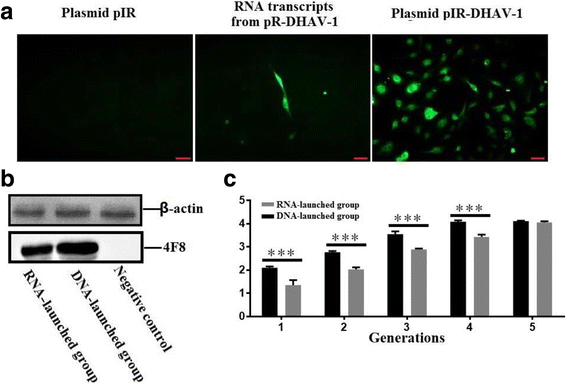
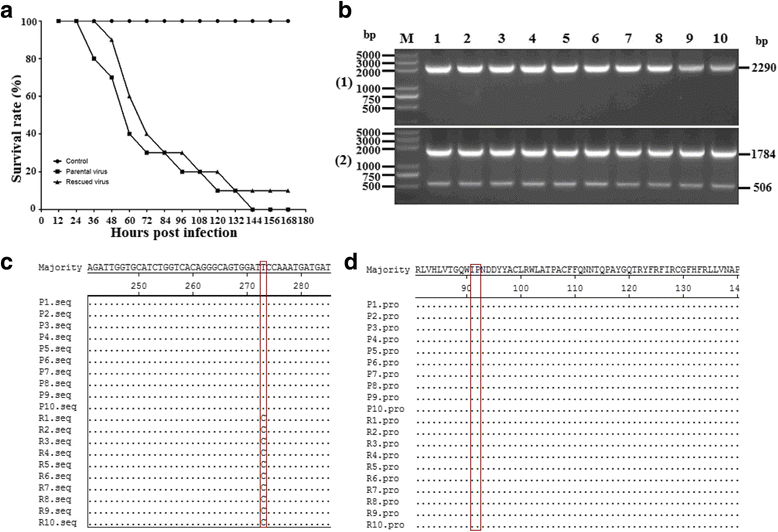
Methods: A total of four overlapping fragments covering the entire viral genome were amplified and then were assembled into a transformation vector based on pIRES2-EGFP to establish the DNA-launched infectious system of duck hepatitis A virus type 1 (DHAV-1), named pIR-DHAV-1. Reverse transcription polymerase chain reaction (RT-PCR) detection, quantitative real-time polymerase chain reaction (qRT-PCR), western blotting assay and indirect immunofluorescence (IFA) were conducted for rescued virus identification. A total of 4.0 μg of recombinant plasmid of pIR-DHAV-1 and in vitro transcribed product of 4.0 μg of RNA-launched infectious clone named pR-DHAV-1 were transfected into BHK-21 cells to analyze the rescue efficiency. Following that, tissue tropism of rescued virus (rDHAV-1) and parental virus (pDHAV-1) were assayed for virulence testing in 1-day-old ducklings.
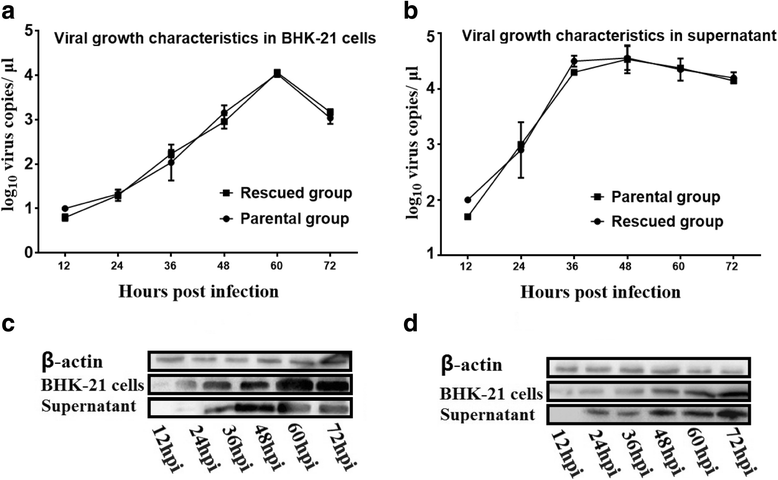
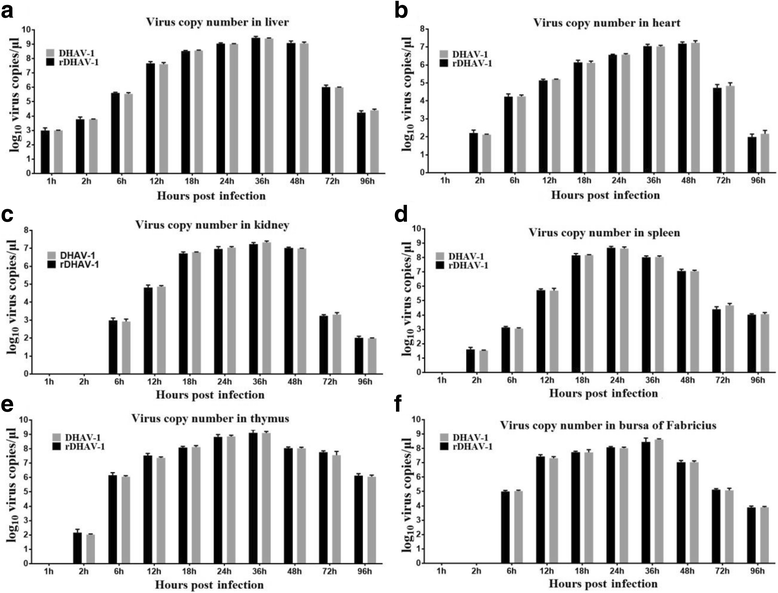
Results: Rescued virus particles carry the designed genetic marker which could be harvested by directly transfecting pIR-DHAV-1 to BHK-21 cells. The qRT-PCR and western blotting results indicated that rDHAV-1 shared similar growth characteristics with pDHAV-1. Furthermore, DNA-launched infectious system possessed much higher rescue efficiency assay compared to RNA-launched infectious system. The mutation at position 3042 from T to C has no impact on viral replication and tissue tropism. From 1 h post infection (hpi) to 48 hpi, the viral RNA copies of rDHAV-1 in liver were the highest among the six tested tissues (with an exception of thymus at 6 hpi), while the viral RNA copy numbers in heart and kidney were alternately the lowest.
Conclusion: We have constructed a genetically stable and highly pathogenic DNA-launched infectious clone, from which the rescued virus could be harvested by direct transfection with recombinant plasmids. rDHAV-1 shared similar growth characteristics and tissue tropism with pDHAV-1. The DNA-launched infectious system of DHAV-1 possessed higher rescue efficiency compared to the traditional RNA-launched infectious system.
Keywords: DHAV-1; DNA-launched infectious clone; Rescue efficiency; Ribozyme.
Conflict of interest statement
Ethics approval
This research was conducted in strict accordance with the recommendations in the Guide for the Institutional Animal Care and Use Commission (IACUC). The animal experiments were carried out in accordance with the guidelines issued by Shandong Agricultural University Animal Care and Use Committee (SDAUA-2014-014). All animal experiments were performed under anesthesia, and every effort was made to minimize suffering.
Consent for publication
Not applicable.
Competing interests
The author(s) declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
A novel method to rescue and culture duck Astrovirus type 1 in vitro.Virol J. 2019 Sep 5;16(1):112. doi: 10.1186/s12985-019-1218-5. Virol J. 2019. PMID: 31488178 Free PMC article.
-
Recovery of duck hepatitis A virus 3 from a stable full-length infectious cDNA clone.Virus Res. 2011 Sep;160(1-2):439-43. doi: 10.1016/j.virusres.2011.07.008. Epub 2011 Jul 27. Virus Res. 2011. PMID: 21801768
-
A one-step duplex rRT-PCR assay for the simultaneous detection of duck hepatitis A virus genotypes 1 and 3.J Virol Methods. 2016 Oct;236:207-214. doi: 10.1016/j.jviromet.2016.07.011. Epub 2016 Jul 18. J Virol Methods. 2016. PMID: 27435338
-
[Overview on duck virus hepatitis A].Sheng Wu Gong Cheng Xue Bao. 2012 Jul;28(7):789-99. Sheng Wu Gong Cheng Xue Bao. 2012. PMID: 23167191 Review. Chinese.
-
Current status and future direction of duck hepatitis A virus vaccines.Avian Pathol. 2023 Apr;52(2):89-99. doi: 10.1080/03079457.2022.2162367. Epub 2023 Jan 19. Avian Pathol. 2023. PMID: 36571394 Review.
Cited by
-
IGF2BP1 Significantly Enhances Translation Efficiency of Duck Hepatitis A Virus Type 1 without Affecting Viral Replication.Biomolecules. 2019 Oct 10;9(10):594. doi: 10.3390/biom9100594. Biomolecules. 2019. PMID: 31658691 Free PMC article.
-
Rapid Rescue of Goose Astrovirus Genome via Red/ET Assembly.Food Environ Virol. 2024 Sep;16(3):297-306. doi: 10.1007/s12560-024-09593-4. Epub 2024 Apr 6. Food Environ Virol. 2024. PMID: 38582780
-
A novel method to rescue and culture duck Astrovirus type 1 in vitro.Virol J. 2019 Sep 5;16(1):112. doi: 10.1186/s12985-019-1218-5. Virol J. 2019. PMID: 31488178 Free PMC article.
-
Interleukin-2 enhancer binding factor 2 negatively regulates the replication of duck hepatitis A virus type 1 by disrupting the RNA-dependent RNA polymerase activity of 3D polymerase.Vet Res. 2024 Mar 26;55(1):40. doi: 10.1186/s13567-024-01294-x. Vet Res. 2024. PMID: 38532469 Free PMC article.
-
The Functional Role of the 3' Untranslated Region and Poly(A) Tail of Duck Hepatitis A Virus Type 1 in Viral Replication and Regulation of IRES-Mediated Translation.Front Microbiol. 2018 Sep 25;9:2250. doi: 10.3389/fmicb.2018.02250. eCollection 2018. Front Microbiol. 2018. PMID: 30319572 Free PMC article.
References
-
- Levine PP, Fabricant JA. Hitherto-undescribed virus disease of ducks in North America. Cornell Vet. 1950;40:71–86.
-
- Woolcock PR. Duck hepatitis. In: Salf YM, Barnes HJ, Glisson JR, Fadly AM, McDougald LR, Swayne DE (Eds) Diseases of poultry, 11th ed, Iowa state press, Ames, IA, 2003;pp. 343–354.
-
- Bosch A, Guix S, Krishna NK, Méndez E, Monroe SS, Pantin-Jackwood M, Schultz-Cherry S. Astroviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus taxonomy. Classification and nomenclature of viruses: ninth report of the international committee on the taxonomy of viruses. Elsevier. London, UK: Academic Press; 2011. pp. 953–959.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

