NMR-directed design of pre-TCRβ and pMHC molecules implies a distinct geometry for pre-TCR relative to αβTCR recognition of pMHC
- PMID: 29101227
- PMCID: PMC5777251
- DOI: 10.1074/jbc.M117.813493
NMR-directed design of pre-TCRβ and pMHC molecules implies a distinct geometry for pre-TCR relative to αβTCR recognition of pMHC
Abstract
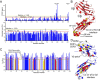
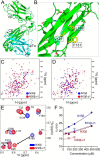
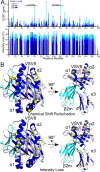
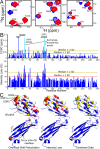
The pre-T cell receptor (pre-TCR) guides early thymocytes through maturation processes within the thymus via interaction with self-ligands displayed on thymic epithelial cells. The pre-TCR is a disulfide-linked heterodimer composed of an invariant pre-TCR α (pTα) subunit and a variable β subunit, the latter of which is incorporated into the mature TCR in subsequent developmental progression. This interaction of pre-TCR with peptide-major histocompatibility complex (pMHC) molecules has recently been shown to drive robust pre-TCR signaling and thymocyte maturation. Although the native sequences of β are properly folded and suitable for NMR studies in isolation, a tendency to self-associate rendered binding studies with physiological ligands difficult to interpret. Consequently, to structurally define this critical interaction, we have re-engineered the extracellular regions of β, designated as β-c1, for prokaryotic production to be used in NMR spectroscopy. Given the large size of the full extracellular domain of class I MHC molecules such as H-Kb, we produced a truncated form termed Kb-t harboring properties favorable for NMR measurements. This system has enabled robust measurement of a pre-TCR-pMHC interaction directly analogous to that of TCRαβ-pMHC. Binding surface analysis identified a contact surface comparable in size to that of the TCRαβ-pMHC but potentially with a rather distinct binding orientation. A tilting of the pre-TCRβ when bound to the pMHC ligand recognition surface versus the upright orientation of TCRαβ would alter the direction of force application between pre-TCR and TCR mechanosensors, impacting signal initiation.
Keywords: T-cell receptor (TCR); immunology; major histocompatibility complex (MHC); nuclear magnetic resonance (NMR); protein domain; protein folding.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures






References
-
- Das D. K., Feng Y., Mallis R. J., Li X., Keskin D. B., Hussey R. E., Brady S. K., Wang J.-H., Wagner G., Reinherz E. L., and Lang M. J. (2015) Force-dependent transition in the T-cell receptor β-subunit allosterically regulates peptide discrimination and pMHC bond lifetime. Proc. Natl. Acad. Sci. U.S.A. 112, 1517–1522 10.1073/pnas.1424829112 - DOI - PMC - PubMed
-
- Liu Y., Blanchfield L., Ma V. P., Andargachew R., Galior K., Liu Z., Evavold B., and Salaita K. (2016) DNA-based nanoparticle tension sensors reveal that T-cell receptors transmit defined pN forces to their antigens for enhanced fidelity. Proc. Natl. Acad. Sci. U.S.A. 113, 5610–5615 10.1073/pnas.1600163113 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

